Identification and Classification for the Lactobacillus casei Group
- PMID: 30186277
- PMCID: PMC6113361
- DOI: 10.3389/fmicb.2018.01974
Identification and Classification for the Lactobacillus casei Group
Abstract
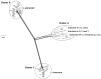
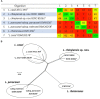
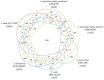
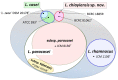
Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus are phenotypically and genotypically closely related, and together comprise the L. casei group. Although the strains of this group are commercially valuable as probiotics, the taxonomic status and nomenclature of the L. casei group have long been contentious because of the difficulties in identifying these three species by using the most frequently used genotypic methodology of 16S rRNA gene sequencing. Long used as the gold standard for species classification, DNA-DNA hybridization is laborious, requires expert skills, and is difficult to use routinely in laboratories. Currently, genome-based comparisons, including average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH), are commonly applied to bacterial taxonomy as alternatives to the gold standard method for the demarcating phylogenetic relationships. To establish quick and accurate methods for identifying strains in the L. casei group at the species and subspecies levels, we developed species- and subspecies-specific identification methods based on housekeeping gene sequences and whole-cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) spectral pattern analysis. By phylogenetic analysis based on concatenated housekeeping gene sequences (dnaJ, dnaK, mutL, pheS, and yycH), 53 strains were separated into four clusters corresponding to the four species: L. casei, L. paracasei and L. rhamnosus, and Lactobacillus chiayiensis sp. nov. A multiplex minisequencing assay using single nucleotide polymorphism (SNP)-specific primers based on the dnaK gene sequences and species-specific primers based on the mutL gene sequences provided high resolution that enabled the strains at the species level to be identified as L. casei, L. paracasei, and L. rhamnosus. By MALDI-TOF MS analysis coupled with an internal database and ClinProTools software, species- and subspecies-level L. casei group strains were identified based on reliable scores and species- and subspecies-specific MS peaks. The L. paracasei strains were distinguished clearly at the subspecies level based on subspecies-specific MS peaks. This article describes the rapid and accurate methods used for identification and classification of strains in the L. casei group based on housekeeping gene sequences and MALDI-TOF MS analysis as well as the novel speciation of this group including L. chiayiensis sp. nov. and 'Lactobacillus zeae' by genome-based methods.
Keywords: Lactobacillus casei group; MALDI-TOF MS; MLSA; species-specific PCR; whole genome sequence.
Figures

Similar articles
-
Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools.J Dairy Sci. 2018 Feb;101(2):979-991. doi: 10.3168/jds.2017-13642. Epub 2017 Nov 15. J Dairy Sci. 2018. PMID: 29153517
-
Polyphasic characterization of a novel species in the Lactobacillus casei group from cow manure of Taiwan: Description of L. chiayiensis sp. nov.Syst Appl Microbiol. 2018 Jul;41(4):270-278. doi: 10.1016/j.syapm.2018.01.008. Epub 2018 Feb 7. Syst Appl Microbiol. 2018. PMID: 29456028
-
Genome-based reclassification of Lactobacillus casei: emended classification and description of the species Lactobacillus zeae.Int J Syst Evol Microbiol. 2020 Jun;70(6):3755-3762. doi: 10.1099/ijsem.0.003969. Int J Syst Evol Microbiol. 2020. PMID: 32421490
-
The Lactobacillus casei Group: History and Health Related Applications.Front Microbiol. 2018 Sep 10;9:2107. doi: 10.3389/fmicb.2018.02107. eCollection 2018. Front Microbiol. 2018. PMID: 30298055 Free PMC article. Review.
-
Multilocus genotyping for classification and genetic structuring of Lactobacillus casei: insights from source and geographical origin.World J Microbiol Biotechnol. 2024 Feb 13;40(3):93. doi: 10.1007/s11274-024-03898-z. World J Microbiol Biotechnol. 2024. PMID: 38349588 Review.
Cited by
-
Evaluating the Probiotic Profile, Antioxidant Properties, and Safety of Indigenous Lactobacillus spp. Inhabiting Fermented Green Tender Coconut Water.Probiotics Antimicrob Proteins. 2024 Sep 20. doi: 10.1007/s12602-024-10352-x. Online ahead of print. Probiotics Antimicrob Proteins. 2024. PMID: 39300004
-
The Skin Histopathology of Pro- and Parabiotics in a Mouse Model of Atopic Dermatitis.Nutrients. 2024 Aug 30;16(17):2903. doi: 10.3390/nu16172903. Nutrients. 2024. PMID: 39275219 Free PMC article.
-
Characterization of probiotics isolated from dietary supplements and evaluation of metabiotic-antibiotic combinations as promising therapeutic options against antibiotic-resistant pathogens using time-kill assay.BMC Complement Med Ther. 2024 Aug 14;24(1):303. doi: 10.1186/s12906-024-04582-3. BMC Complement Med Ther. 2024. PMID: 39143578 Free PMC article.
-
Lacticaseibacillus parahuelsenbergensis sp. nov., Lacticaseibacillus styriensis sp. nov. and Lacticaseibacillus zeae subsp. silagei subsp. nov., isolated from different grass and corn silage.Int J Syst Evol Microbiol. 2024 Jul;74(7):006441. doi: 10.1099/ijsem.0.006441. Int J Syst Evol Microbiol. 2024. PMID: 38954457
-
Antibacterial potential of lactic acid bacteria isolated from raw cow milk in Sylhet district, Bangladesh: A molecular approach.Vet Med Sci. 2024 May;10(3):e1463. doi: 10.1002/vms3.1463. Vet Med Sci. 2024. PMID: 38659354 Free PMC article.
References
-
- Ahrné S., Molin G., StÅhl S. (1989). Plasmids in Lactobacillus strains isolated from meat and meat products. Syst. Appl. Microbiol. 11 320–325. 10.1016/S0723-2020(89)80031-1 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources


