Hepatitis A Virus Capsid Structure
- PMID: 30037986
- PMCID: PMC6496327
- DOI: 10.1101/cshperspect.a031807
Hepatitis A Virus Capsid Structure
Abstract
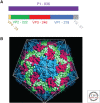
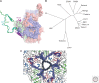
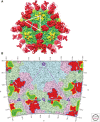
Hepatitis A virus (HAV) has been enigmatic, evading detailed structural analysis for many years. Its recently determined high-resolution structure revealed an angular surface without the indentations often characteristic of receptor-binding sites. The viral protein 1 (VP1) β-barrel shows no sign of a pocket factor and the amino terminus of VP2 displays a "domain swap" across the pentamer interface, as in a subset of mammalian picornaviruses and insect picorna-like viruses. Structure-based phylogeny confirms this placement. These differences suggest an uncoating mechanism distinct from that of enteroviruses. An empty capsid structure reveals internal differences in VP0 and the VP1 amino terminus connected with particle maturation. An HAV/Fab complex structure, in which the antigen-binding fragment (Fab) appears to act as a receptor-mimic, clarifies some historical epitope mapping data, but some remain difficult to reconcile. We still have little idea of the structural features of enveloped HAV particles.
Copyright © 2019 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


Similar articles
-
Hepatitis A virus and the origins of picornaviruses.Nature. 2015 Jan 1;517(7532):85-88. doi: 10.1038/nature13806. Epub 2014 Oct 19. Nature. 2015. PMID: 25327248 Free PMC article.
-
Redundant Late Domain Functions of Tandem VP2 YPX3L Motifs in Nonlytic Cellular Egress of Quasi-enveloped Hepatitis A Virus.J Virol. 2018 Nov 12;92(23):e01308-18. doi: 10.1128/JVI.01308-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232181 Free PMC article.
-
Virion Structure of Israeli Acute Bee Paralysis Virus.J Virol. 2016 Aug 26;90(18):8150-9. doi: 10.1128/JVI.00854-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384649 Free PMC article.
-
Hepatitis A Virus Genome Organization and Replication Strategy.Cold Spring Harb Perspect Med. 2018 Dec 3;8(12):a033480. doi: 10.1101/cshperspect.a033480. Cold Spring Harb Perspect Med. 2018. PMID: 29610147 Free PMC article. Review.
-
Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation.Curr Opin Virol. 2016 Jun;18:82-8. doi: 10.1016/j.coviro.2016.05.002. Epub 2016 May 30. Curr Opin Virol. 2016. PMID: 27253691 Review.
Cited by
-
A Useful Method to Provide Infectious and Cultivable In Vitro Naked Viral Particles of Hepatitis A Virus.Viruses. 2024 Aug 26;16(9):1360. doi: 10.3390/v16091360. Viruses. 2024. PMID: 39339837 Free PMC article.
-
Inactivation of Hepatitis A Virus and Feline Calicivirus on Model Food Contact Surfaces by Ultraviolet Light (UV-C) Systems.Foods. 2024 Sep 12;13(18):2892. doi: 10.3390/foods13182892. Foods. 2024. PMID: 39335821 Free PMC article.
-
Cellular immune response to a single dose of live attenuated hepatitis a virus vaccine in obese children and adolescents.Heliyon. 2024 Aug 20;10(16):e36610. doi: 10.1016/j.heliyon.2024.e36610. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39258209 Free PMC article.
-
Identification and Genomic Characterization of Two Novel Hepatoviruses in Shrews from Yunnan Province, China.Viruses. 2024 Jun 17;16(6):969. doi: 10.3390/v16060969. Viruses. 2024. PMID: 38932262 Free PMC article.
-
Genomic characterization of a novel Hepatovirus identified in Maranhão state, Brazil.Sci Rep. 2024 Apr 5;14(1):7981. doi: 10.1038/s41598-024-58171-y. Sci Rep. 2024. PMID: 38575654 Free PMC article.
References
-
- Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature 337: 709–716. - PubMed
-
- Chapman MS, Rossmann MG. 1993. Comparison of surface properties of picornaviruses: Strategies for hiding the receptor site from immune surveillance. Virology 195: 745–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

