Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria
- PMID: 29666938
- PMCID: PMC6133090
- DOI: 10.1007/s00709-018-1241-1
Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria
Abstract
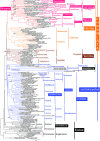
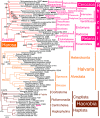
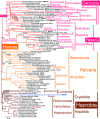
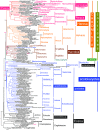
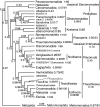
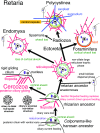
Infrakingdom Rhizaria is one of four major subgroups with distinct cell body plans that comprise eukaryotic kingdom Chromista. Unlike other chromists, Rhizaria are mostly heterotrophic flagellates, amoebae or amoeboflagellates, commonly with reticulose (net-like) or filose (thread-like) feeding pseudopodia; uniquely for eukaryotes, cilia have proximal ciliary transition-zone hub-lattices. They comprise predominantly flagellate phylum Cercozoa and reticulopodial phylum Retaria, whose exact phylogenetic relationship has been uncertain. Given even less clear relationships amongst cercozoan classes, we sequenced partial transcriptomes of seven Cercozoa representing five classes and endomyxan retarian Filoreta marina to establish 187-gene multiprotein phylogenies. Ectoreta (retarian infraphyla Foraminifera, Radiozoa) branch within classical Cercozoa as sister to reticulose Endomyxa. This supports recent transfer of subphylum Endomyxa from Cercozoa to Retaria alongside subphylum Ectoreta which embraces classical retarians where capsules or tests subdivide cells into organelle-containing endoplasm and anastomosing pseudopodial net-like ectoplasm. Cercozoa are more homogeneously filose, often with filose pseudopodia and/or posterior ciliary gliding motility: zooflagellate Helkesimastix and amoeboid Guttulinopsis form a strongly supported clade, order Helkesida. Cercomonads are polyphyletic (Cercomonadida sister to glissomonads; Paracercomonadida deeper). Thecofilosea are a clade, whereas Imbricatea may not be; Sarcomonadea may be paraphyletic. Helkesea and Metromonadea are successively deeper outgroups within cercozoan subphylum Monadofilosa; subphylum Reticulofilosa (paraphyletic on site-heterogeneous trees) branches earliest, Granofilosea before Chlorarachnea. Our multiprotein trees confirm that Rhizaria are sisters of infrakingdom Halvaria (Alveolata, Heterokonta) within chromist subkingdom Harosa (= SAR); they further support holophyly of chromist subkingdom Hacrobia, and are consistent with holophyly of Chromista as sister of kingdom Plantae. Site-heterogeneous rDNA trees group Kraken with environmental DNA clade 'eSarcomonad', not Paracercomonadida. Ectoretan fossil dates evidence ultrarapid episodic stem sequence evolution. We discuss early rhizarian cell evolution and multigene tree coevolutionary patterns, gene-paralogue evidence for chromist monophyly, and integrate this with fossil evidence for the age of Rhizaria and eukaryote cells, and revise rhizarian classification.
Keywords: Cell evolution; Cercozoa; Chromista; Harosa; Retaria; Rhizarian phylogeny.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures

Similar articles
-
Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution.J Mol Evol. 2003 May;56(5):540-63. doi: 10.1007/s00239-002-2424-z. J Mol Evol. 2003. PMID: 12698292
-
Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences.Protoplasma. 2018 Jan;255(1):297-357. doi: 10.1007/s00709-017-1147-3. Epub 2017 Sep 5. Protoplasma. 2018. PMID: 28875267 Free PMC article.
-
Evolutionary Relationship of the Scale-Bearing Kraken (incertae sedis, Monadofilosa, Cercozoa, Rhizaria): Combining Ultrastructure Data and a Two-Gene Phylogeny.Protist. 2017 Jul;168(3):362-373. doi: 10.1016/j.protis.2017.04.004. Epub 2017 May 10. Protist. 2017. PMID: 28582680
-
The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa.Int J Syst Evol Microbiol. 2002 Mar;52(Pt 2):297-354. doi: 10.1099/00207713-52-2-297. Int J Syst Evol Microbiol. 2002. PMID: 11931142 Review.
-
Ciliary transition zone evolution and the root of the eukaryote tree: implications for opisthokont origin and classification of kingdoms Protozoa, Plantae, and Fungi.Protoplasma. 2022 May;259(3):487-593. doi: 10.1007/s00709-021-01665-7. Epub 2021 Dec 23. Protoplasma. 2022. PMID: 34940909 Free PMC article. Review.
Cited by
-
Fine-scale characterization of the soybean rhizosphere microbiome via synthetic long reads and avidity sequencing.Environ Microbiome. 2024 Jul 12;19(1):46. doi: 10.1186/s40793-024-00590-5. Environ Microbiome. 2024. PMID: 38997772 Free PMC article.
-
Comparative genomics of Ascetosporea gives new insight into the evolutionary basis for animal parasitism in Rhizaria.BMC Biol. 2024 May 3;22(1):103. doi: 10.1186/s12915-024-01898-x. BMC Biol. 2024. PMID: 38702750 Free PMC article.
-
Transition from stromatolite to thrombolite fabric: potential role for reticulopodial protists in lake microbialites of a Proterozoic ecosystem analog.Front Microbiol. 2023 Oct 30;14:1210781. doi: 10.3389/fmicb.2023.1210781. eCollection 2023. Front Microbiol. 2023. PMID: 37965561 Free PMC article.
-
The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections.Microorganisms. 2023 Jan 23;11(2):295. doi: 10.3390/microorganisms11020295. Microorganisms. 2023. PMID: 36838260 Free PMC article. Review.
-
Phagocytosis underpins the biotrophic lifestyle of intracellular parasites in the class Phytomyxea (Rhizaria).New Phytol. 2023 Jun;238(5):2130-2143. doi: 10.1111/nph.18828. Epub 2023 Mar 24. New Phytol. 2023. PMID: 36810975 Free PMC article.
References
-
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, le Gall L, Lynn DH, McManus H, Mitchell EAD, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. - PMC - PubMed
-
- Allen CA, Jackson AP, Rigden DJ, Willis AC, Ferguson AC, Ginger ML. Order within a mosaic distribution of mitochondrial c-type cytochrome biogenesis systems? FEBS J. 2008;275:2385–2402. - PubMed
-
- Antcliffe JB. Questioning the evidence of organic compounds called sponge biomarkers. Palaeontology. 2013;56:917–925.
-
- Arnold ZM. Observations on the sexual generation of Gromia oviformis Dujardin. J Protozool. 1966;13:23–27. - PubMed
-
- Arnold ZM. Observations on the biology of the protozoan Gromia oviformis Dujardin. Univ Calif Publ Zool. 1972;100:1–168.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous


