Reconstructing the genetic history of late Neanderthals
- PMID: 29562232
- PMCID: PMC6485383
- DOI: 10.1038/nature26151
Reconstructing the genetic history of late Neanderthals
Abstract
Although it has previously been shown that Neanderthals contributed DNA to modern humans, not much is known about the genetic diversity of Neanderthals or the relationship between late Neanderthal populations at the time at which their last interactions with early modern humans occurred and before they eventually disappeared. Our ability to retrieve DNA from a larger number of Neanderthal individuals has been limited by poor preservation of endogenous DNA and contamination of Neanderthal skeletal remains by large amounts of microbial and present-day human DNA. Here we use hypochlorite treatment of as little as 9 mg of bone or tooth powder to generate between 1- and 2.7-fold genomic coverage of five Neanderthals who lived around 39,000 to 47,000 years ago (that is, late Neanderthals), thereby doubling the number of Neanderthals for which genome sequences are available. Genetic similarity among late Neanderthals is well predicted by their geographical location, and comparison to the genome of an older Neanderthal from the Caucasus indicates that a population turnover is likely to have occurred, either in the Caucasus or throughout Europe, towards the end of Neanderthal history. We find that the bulk of Neanderthal gene flow into early modern humans originated from one or more source populations that diverged from the Neanderthals that were studied here at least 70,000 years ago, but after they split from a previously sequenced Neanderthal from Siberia around 150,000 years ago. Although four of the Neanderthals studied here post-date the putative arrival of early modern humans into Europe, we do not detect any recent gene flow from early modern humans in their ancestry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
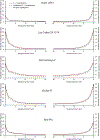
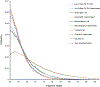



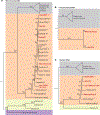

Similar articles
-
The complete genome sequence of a Neanderthal from the Altai Mountains.Nature. 2014 Jan 2;505(7481):43-9. doi: 10.1038/nature12886. Epub 2013 Dec 18. Nature. 2014. PMID: 24352235 Free PMC article.
-
Ancient gene flow from early modern humans into Eastern Neanderthals.Nature. 2016 Feb 25;530(7591):429-33. doi: 10.1038/nature16544. Epub 2016 Feb 17. Nature. 2016. PMID: 26886800 Free PMC article.
-
Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry.Nature. 2021 Apr;592(7853):253-257. doi: 10.1038/s41586-021-03335-3. Epub 2021 Apr 7. Nature. 2021. PMID: 33828320 Free PMC article.
-
Archaic human genomics.Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review.
-
Archaic admixture in human history.Curr Opin Genet Dev. 2016 Dec;41:93-97. doi: 10.1016/j.gde.2016.07.002. Epub 2016 Sep 20. Curr Opin Genet Dev. 2016. PMID: 27662059 Review.
Cited by
-
Initial Upper Palaeolithic lithic industry at Cueva Millán in the hinterlands of Iberia.Sci Rep. 2024 Sep 27;14(1):21705. doi: 10.1038/s41598-024-69913-3. Sci Rep. 2024. PMID: 39333171 Free PMC article.
-
Long genetic and social isolation in Neanderthals before their extinction.Cell Genom. 2024 Sep 11;4(9):100593. doi: 10.1016/j.xgen.2024.100593. Cell Genom. 2024. PMID: 39265525 Free PMC article.
-
Reconstructing contact and a potential interbreeding geographical zone between Neanderthals and anatomically modern humans.Sci Rep. 2024 Sep 3;14(1):20475. doi: 10.1038/s41598-024-70206-y. Sci Rep. 2024. PMID: 39227643 Free PMC article.
-
Swordtail fish hybrids reveal that genome evolution is surprisingly predictable after initial hybridization.PLoS Biol. 2024 Aug 26;22(8):e3002742. doi: 10.1371/journal.pbio.3002742. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39186811 Free PMC article.
-
A disease-associated gene desert directs macrophage inflammation through ETS2.Nature. 2024 Jun;630(8016):447-456. doi: 10.1038/s41586-024-07501-1. Epub 2024 Jun 5. Nature. 2024. PMID: 38839969 Free PMC article.
References
-
- Pääbo S et al. Genetic analyses from ancient DNA. Annu. Rev. Genet 38, 645–679 (2004). - PubMed
-
- Gilbert MT, Bandelt HJ, Hofreiter M & Barnes I Assessing ancient DNA studies. Trends Ecol. Evol 20, 541–544 (2005). - PubMed
-
- Krause J et al. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol 20, 231–236 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


