Site-Specific Disulfide Crosslinked Nucleosomes with Enhanced Stability
- PMID: 29113904
- PMCID: PMC5757783
- DOI: 10.1016/j.jmb.2017.10.029
Site-Specific Disulfide Crosslinked Nucleosomes with Enhanced Stability
Abstract
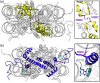
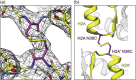
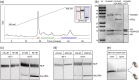
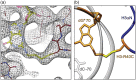
We engineered nucleosome core particles (NCPs) with two site-specific cysteine crosslinks that increase the stability of the particle. The first disulfide was introduced between the two copies of H2A via an H2A-N38C point mutation, effectively crosslinking the two H2A/H2B heterodimers together to stabilize the histone octamer against H2A/H2B dimer dissociation. The second crosslink was engineered between an R40C point mutation on the N-terminal tail of H3 and the NCP DNA ends by the introduction of a convertible nucleotide. This crosslink maintains the nucleosome DNA in a fixed translational setting relative to the histone octamer and prevents dilution-driven dissociation. The X-ray crystal structures of NCPs containing the disulfides in isolation and in combination were determined. Both disulfides stabilize the structure of the NCP without disturbing the overall structure. Nucleosomes containing these modifications will be advantageous for biochemical and structural studies as a consequence of their greater resistance to dissociation during high dilution in purification, elevated salt for crystallization and vitrification for cryogenic electron microscopy.
Keywords: DNA; X-ray crystallography; chromatin; hexasome; histone.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

Similar articles
-
Protein-protein Förster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z.J Mol Biol. 2007 Aug 24;371(4):971-88. doi: 10.1016/j.jmb.2007.05.075. Epub 2007 Jun 2. J Mol Biol. 2007. PMID: 17597150 Free PMC article.
-
The histone variant macro-H2A preferentially forms "hybrid nucleosomes".J Biol Chem. 2006 Sep 1;281(35):25522-31. doi: 10.1074/jbc.M602258200. Epub 2006 Jun 27. J Biol Chem. 2006. PMID: 16803903
-
H2A and H2B tails are essential to properly reconstitute nucleosome core particles.Eur Biophys J. 2007 Nov;36(8):1083-94. doi: 10.1007/s00249-007-0212-9. Epub 2007 Sep 19. Eur Biophys J. 2007. PMID: 17882413
-
The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective.Biochim Biophys Acta. 2009 Jan;1789(1):37-44. doi: 10.1016/j.bbagrm.2008.07.001. Epub 2008 Jul 14. Biochim Biophys Acta. 2009. PMID: 18675384 Review.
-
Patterning chromatin: form and function for H2A.Z variant nucleosomes.Curr Opin Genet Dev. 2006 Apr;16(2):119-24. doi: 10.1016/j.gde.2006.02.005. Epub 2006 Feb 28. Curr Opin Genet Dev. 2006. PMID: 16503125 Review.
Cited by
-
Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.Science. 2023 Jul 21;381(6655):319-324. doi: 10.1126/science.adf4197. Epub 2023 Jun 29. Science. 2023. PMID: 37384669 Free PMC article.
-
Expressed Protein Ligation without Intein.J Am Chem Soc. 2020 Apr 15;142(15):7047-7054. doi: 10.1021/jacs.0c00252. Epub 2020 Apr 2. J Am Chem Soc. 2020. PMID: 32212692 Free PMC article.
-
Molecular engineering strategies for visualizing low-affinity protein complexes.Exp Biol Med (Maywood). 2019 Dec;244(17):1559-1567. doi: 10.1177/1535370219855401. Epub 2019 Jun 11. Exp Biol Med (Maywood). 2019. PMID: 31184923 Free PMC article.
-
Automated cryo-EM structure refinement using correlation-driven molecular dynamics.Elife. 2019 Mar 4;8:e43542. doi: 10.7554/eLife.43542. Elife. 2019. PMID: 30829573 Free PMC article.
-
Bend-Induced Twist Waves and the Structure of Nucleosomal DNA.Phys Rev Lett. 2018 Aug 24;121(8):088101. doi: 10.1103/PhysRevLett.121.088101. Phys Rev Lett. 2018. PMID: 30192578 Free PMC article.
References
-
- Kornberg R. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. - PubMed
-
- van Holde K. Springer; New York: 1989. Chromatin.
-
- Luger K., Maeder A., Richmond R., Sargent D., Richmond T. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Luger K., Rechsteiner T., Flaus A., Waye M., Richmond T. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


