PULDB: the expanded database of Polysaccharide Utilization Loci
- PMID: 29088389
- PMCID: PMC5753385
- DOI: 10.1093/nar/gkx1022
PULDB: the expanded database of Polysaccharide Utilization Loci
Abstract
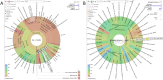
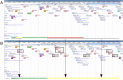
The Polysaccharide Utilization Loci (PUL) database was launched in 2015 to present PUL predictions in ∼70 Bacteroidetes species isolated from the human gastrointestinal tract, as well as PULs derived from the experimental data reported in the literature. In 2018 PULDB offers access to 820 genomes, sampled from various environments and covering a much wider taxonomical range. A Krona dynamic chart was set up to facilitate browsing through taxonomy. Literature surveys now allows the presentation of the most recent (i) PUL repertoires deduced from RNAseq large-scale experiments, (ii) PULs that have been subjected to in-depth biochemical analysis and (iii) new Carbohydrate-Active enzyme (CAZyme) families that contributed to the refinement of PUL predictions. To improve PUL visualization and genome browsing, the previous annotation of genes encoding CAZymes, regulators, integrases and SusCD has now been expanded to include functionally relevant protein families whose genes are significantly found in the vicinity of PULs: sulfatases, proteases, ROK repressors, epimerases and ATP-Binding Cassette and Major Facilitator Superfamily transporters. To cope with cases where susCD may be absent due to incomplete assemblies/split PULs, we present 'CAZyme cluster' predictions. Finally, a PUL alignment tool, operating on the tagged families instead of amino-acid sequences, was integrated to retrieve PULs similar to a query of interest. The updated PULDB website is accessible at www.cazy.org/PULDB_new/.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
dbCAN-PUL: a database of experimentally characterized CAZyme gene clusters and their substrates.Nucleic Acids Res. 2021 Jan 8;49(D1):D523-D528. doi: 10.1093/nar/gkaa742. Nucleic Acids Res. 2021. PMID: 32941621 Free PMC article.
-
Automatic prediction of polysaccharide utilization loci in Bacteroidetes species.Bioinformatics. 2015 Mar 1;31(5):647-55. doi: 10.1093/bioinformatics/btu716. Epub 2014 Oct 28. Bioinformatics. 2015. PMID: 25355788
-
Polysaccharide Utilization Loci: Fueling Microbial Communities.J Bacteriol. 2017 Jul 11;199(15):e00860-16. doi: 10.1128/JB.00860-16. Print 2017 Aug 1. J Bacteriol. 2017. PMID: 28138099 Free PMC article. Review.
-
Bacteroidetes use thousands of enzyme combinations to break down glycans.Nat Commun. 2019 May 3;10(1):2043. doi: 10.1038/s41467-019-10068-5. Nat Commun. 2019. PMID: 31053724 Free PMC article.
-
Polysaccharide degradation by the Bacteroidetes: mechanisms and nomenclature.Environ Microbiol Rep. 2021 Oct;13(5):559-581. doi: 10.1111/1758-2229.12980. Epub 2021 Jun 13. Environ Microbiol Rep. 2021. PMID: 34036727 Review.
Cited by
-
Identification and characterization of the lipoprotein N-acyltransferase in Bacteroides.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2410909121. doi: 10.1073/pnas.2410909121. Epub 2024 Nov 4. Proc Natl Acad Sci U S A. 2024. PMID: 39495918
-
In vivo manipulation of human gut Bacteroides fitness by abiotic oligosaccharides.Nat Chem Biol. 2024 Oct 23. doi: 10.1038/s41589-024-01763-6. Online ahead of print. Nat Chem Biol. 2024. PMID: 39443715
-
A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary β-glucans.NPJ Biofilms Microbiomes. 2024 Oct 14;10(1):105. doi: 10.1038/s41522-024-00578-6. NPJ Biofilms Microbiomes. 2024. PMID: 39397008 Free PMC article.
-
A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.Nat Commun. 2024 Oct 10;15(1):8794. doi: 10.1038/s41467-024-53149-w. Nat Commun. 2024. PMID: 39389974 Free PMC article.
-
Determinants of raffinose family oligosaccharide use in Bacteroides species.J Bacteriol. 2024 Oct 24;206(10):e0023524. doi: 10.1128/jb.00235-24. Epub 2024 Sep 27. J Bacteriol. 2024. PMID: 39330254 Free PMC article.
References
-
- El Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B.. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013; 11:497–504. - PubMed
-
- Terrapon N., Lombard V., Gilbert H.J., Henrissat B.. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015; 31:647–655. - PubMed
-
- Westover B.P., Buhler J.D., Sonnenburg J.L., Gordon J.I.. Operon prediction without a training set. Bioinformatics. 2005; 21:880–888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


