Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9
- PMID: 28382278
- PMCID: PMC5360708
- DOI: 10.3389/fcimb.2017.00091
Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9
Abstract
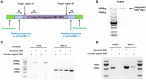
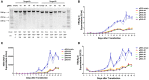
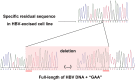
The presence of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) and the permanent integration of HBV DNA into the host genome confers the risk of viral reactivation and hepatocellular carcinoma. Nucleoside/nucleotide analogs alone have little or no capacity to eliminate replicative HBV templates consisting of cccDNA or integrated HBV DNA. Recently, CRISPR/Cas9 technology has been widely applied as a promising genome-editing tool, and HBV-specific CRISPR-Cas9 systems were shown to effectively mediate HBV cccDNA disruption. However, the integrated HBV DNA fragments are considered as important pro-oncogenic properties and it serves as an important template for viral replication and expression in stable HBV cell line. In this study, we completely excised a full-length 3,175-bp integrated HBV DNA fragment and disrupted HBV cccDNA in a stable HBV cell line. In HBV-excised cell line, the HBV cccDNA inside cells, supernatant HBV DNA, HBsAg, and HBeAg remained below the negative critical values for more than 10 months. Besides, by whole genome sequencing, we analyzed off-target effects and excluded cell contamination. It is the first time that the HBV infection has been fully eradicated in a stable HBV cell line. These findings demonstrate that the CRISPR-Cas9 system is a potentially powerful tool capable of promoting a radical or "sterile" HBV cure.
Keywords: CRISPR-Cas9; HBV cccDNA; hepatitis B virus; integrated HBV DNA; whole genome sequencing.
Figures

Similar articles
-
Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication.Antiviral Res. 2015 Jun;118:110-7. doi: 10.1016/j.antiviral.2015.03.015. Epub 2015 Apr 3. Antiviral Res. 2015. PMID: 25843425
-
Application of CRISPR/Cas9 Technology to HBV.Int J Mol Sci. 2015 Nov 2;16(11):26077-86. doi: 10.3390/ijms161125950. Int J Mol Sci. 2015. PMID: 26540039 Free PMC article. Review.
-
CRISPR-Cas9 Targeting of Hepatitis B Virus Covalently Closed Circular DNA Generates Transcriptionally Active Episomal Variants.mBio. 2022 Apr 26;13(2):e0288821. doi: 10.1128/mbio.02888-21. Epub 2022 Apr 7. mBio. 2022. PMID: 35389262 Free PMC article.
-
Synthetic gRNA/Cas9 ribonucleoprotein targeting HBV DNA inhibits viral replication.J Med Virol. 2023 Jul;95(7):e28952. doi: 10.1002/jmv.28952. J Med Virol. 2023. PMID: 37455550 Free PMC article.
-
Novel therapeutic approaches for hepatitis B virus covalently closed circular DNA.World J Gastroenterol. 2015 Jun 21;21(23):7084-8. doi: 10.3748/wjg.v21.i23.7084. World J Gastroenterol. 2015. PMID: 26109795 Free PMC article. Review.
Cited by
-
Prophylactic and therapeutic vaccine development: advancements and challenges.Mol Biomed. 2024 Nov 11;5(1):57. doi: 10.1186/s43556-024-00222-x. Mol Biomed. 2024. PMID: 39527305 Free PMC article. Review.
-
The potential of HBV cure: an overview of CRISPR-mediated HBV gene disruption.Front Genome Ed. 2024 Oct 9;6:1467449. doi: 10.3389/fgeed.2024.1467449. eCollection 2024. Front Genome Ed. 2024. PMID: 39444780 Free PMC article. Review.
-
The impact of integrated hepatitis B virus DNA on oncogenesis and antiviral therapy.Biomark Res. 2024 Aug 15;12(1):84. doi: 10.1186/s40364-024-00611-y. Biomark Res. 2024. PMID: 39148134 Free PMC article. Review.
-
A temperature-sensitive and interferon-silent Sendai virus vector for CRISPR-Cas9 delivery and gene editing in primary human cells.bioRxiv [Preprint]. 2024 May 5:2024.05.03.592383. doi: 10.1101/2024.05.03.592383. bioRxiv. 2024. PMID: 38746439 Free PMC article. Preprint.
-
Harnessing CRISPR technology for viral therapeutics and vaccines: from preclinical studies to clinical applications.Virus Res. 2024 Mar;341:199314. doi: 10.1016/j.virusres.2024.199314. Epub 2024 Jan 12. Virus Res. 2024. PMID: 38211734 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials


