Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score
- PMID: 28323831
- PMCID: PMC5360219
- DOI: 10.1371/journal.pmed.1002258
Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score
Erratum in
-
Correction: Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score.PLoS Med. 2017 Mar 28;14(3):e1002289. doi: 10.1371/journal.pmed.1002289. eCollection 2017 Mar. PLoS Med. 2017. PMID: 28350793 Free PMC article.
Abstract
Background: Identifying individuals at risk for developing Alzheimer disease (AD) is of utmost importance. Although genetic studies have identified AD-associated SNPs in APOE and other genes, genetic information has not been integrated into an epidemiological framework for risk prediction.
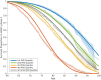
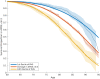
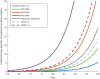
Methods and findings: Using genotype data from 17,008 AD cases and 37,154 controls from the International Genomics of Alzheimer's Project (IGAP Stage 1), we identified AD-associated SNPs (at p < 10-5). We then integrated these AD-associated SNPs into a Cox proportional hazard model using genotype data from a subset of 6,409 AD patients and 9,386 older controls from Phase 1 of the Alzheimer's Disease Genetics Consortium (ADGC), providing a polygenic hazard score (PHS) for each participant. By combining population-based incidence rates and the genotype-derived PHS for each individual, we derived estimates of instantaneous risk for developing AD, based on genotype and age, and tested replication in multiple independent cohorts (ADGC Phase 2, National Institute on Aging Alzheimer's Disease Center [NIA ADC], and Alzheimer's Disease Neuroimaging Initiative [ADNI], total n = 20,680). Within the ADGC Phase 1 cohort, individuals in the highest PHS quartile developed AD at a considerably lower age and had the highest yearly AD incidence rate. Among APOE ε3/3 individuals, the PHS modified expected age of AD onset by more than 10 y between the lowest and highest deciles (hazard ratio 3.34, 95% CI 2.62-4.24, p = 1.0 × 10-22). In independent cohorts, the PHS strongly predicted empirical age of AD onset (ADGC Phase 2, r = 0.90, p = 1.1 × 10-26) and longitudinal progression from normal aging to AD (NIA ADC, Cochran-Armitage trend test, p = 1.5 × 10-10), and was associated with neuropathology (NIA ADC, Braak stage of neurofibrillary tangles, p = 3.9 × 10-6, and Consortium to Establish a Registry for Alzheimer's Disease score for neuritic plaques, p = 6.8 × 10-6) and in vivo markers of AD neurodegeneration (ADNI, volume loss within the entorhinal cortex, p = 6.3 × 10-6, and hippocampus, p = 7.9 × 10-5). Additional prospective validation of these results in non-US, non-white, and prospective community-based cohorts is necessary before clinical use.
Conclusions: We have developed a PHS for quantifying individual differences in age-specific genetic risk for AD. Within the cohorts studied here, polygenic architecture plays an important role in modifying AD risk beyond APOE. With thorough validation, quantification of inherited genetic variation may prove useful for stratifying AD risk and as an enrichment strategy in therapeutic trials.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: JBB served on advisory boards for Elan, Bristol-Myers Squibb, Avanir, Novartis, Genentech, and Eli Lilly and holds stock options in CorTechs Labs, Inc. and Human Longevity, Inc. AMD is a founder of and holds equity in CorTechs Labs, Inc., and serves on its Scientific Advisory Board. He is also a member of the Scientific Advisory Board of Human Longevity, Inc. (HLI), and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. AG served on or have served on in the last 3 years the scientific advisory boards of the following companies: Denali Therapeutics, Cognition Therapeutics and AbbVie. BM served as guest editor on PLOS Medicine’s Special Issue on Dementia.
Figures


Comment in
-
Predicting Alzheimer's disease: a polygenic hazard score.J R Coll Physicians Edinb. 2017 Jun;47(2):151-152. doi: 10.4997/JRCPE.2017.211. J R Coll Physicians Edinb. 2017. PMID: 28675188 No abstract available.
Similar articles
-
Polygenic risk and hazard scores for Alzheimer's disease prediction.Ann Clin Transl Neurol. 2019 Feb 18;6(3):456-465. doi: 10.1002/acn3.716. eCollection 2019 Mar. Ann Clin Transl Neurol. 2019. PMID: 30911569 Free PMC article.
-
Polygenic hazard score: an enrichment marker for Alzheimer's associated amyloid and tau deposition.Acta Neuropathol. 2018 Jan;135(1):85-93. doi: 10.1007/s00401-017-1789-4. Epub 2017 Nov 24. Acta Neuropathol. 2018. PMID: 29177679 Free PMC article.
-
Polygenic hazard scores in preclinical Alzheimer disease.Ann Neurol. 2017 Sep;82(3):484-488. doi: 10.1002/ana.25029. Ann Neurol. 2017. PMID: 28940650 Free PMC article.
-
APOE genotype effects on Alzheimer's disease onset and epidemiology.J Mol Neurosci. 2004;23(3):157-65. doi: 10.1385/JMN:23:3:157. J Mol Neurosci. 2004. PMID: 15181244 Review.
-
ApoE genotype accounts for the vast majority of AD risk and AD pathology.Neurobiol Aging. 2004 May-Jun;25(5):641-50. doi: 10.1016/j.neurobiolaging.2003.12.023. Neurobiol Aging. 2004. PMID: 15172743 Review.
Cited by
-
Validation of a Community-Based Approach Toward Personalized Dementia Risk Reduction: The Kimel Family Centre for Brain Health and Wellness.J Prev Alzheimers Dis. 2024;11(5):1455-1466. doi: 10.14283/jpad.2024.98. J Prev Alzheimers Dis. 2024. PMID: 39350393 Free PMC article.
-
Integrating amyloid and tau imaging with proteomics and genomics in Alzheimer's disease.Cell Rep Med. 2024 Sep 17;5(9):101735. doi: 10.1016/j.xcrm.2024.101735. Cell Rep Med. 2024. PMID: 39293391 Free PMC article. Review.
-
Integrating amyloid imaging and genetics for early risk stratification of Alzheimer's disease.Alzheimers Dement. 2024 Nov;20(11):7819-7830. doi: 10.1002/alz.14244. Epub 2024 Sep 17. Alzheimers Dement. 2024. PMID: 39285750 Free PMC article.
-
Predicting CDR status over 36 months with a recall-based digital cognitive biomarker.Alzheimers Dement. 2024 Oct;20(10):7274-7280. doi: 10.1002/alz.14213. Epub 2024 Sep 11. Alzheimers Dement. 2024. PMID: 39258756 Free PMC article.
-
Alzheimer's disease genetic risk and changes in brain atrophy and white matter hyperintensities in cognitively unimpaired adults.Brain Commun. 2024 Aug 14;6(5):fcae276. doi: 10.1093/braincomms/fcae276. eCollection 2024. Brain Commun. 2024. PMID: 39229494 Free PMC article.
References
-
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH100351/MH/NIMH NIH HHS/United States
- MR/L501542/1/MRC_/Medical Research Council/United Kingdom
- R01 AG054076/AG/NIA NIH HHS/United States
- G1001253/MRC_/Medical Research Council/United Kingdom
- P30 AG013846/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- G0901254/MRC_/Medical Research Council/United Kingdom
- K01 AG049152/AG/NIA NIH HHS/United States
- K24 AG045333/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- R01 GM104400/GM/NIGMS NIH HHS/United States
- MR/K01417X/1/MRC_/Medical Research Council/United Kingdom
- G0701075/MRC_/Medical Research Council/United Kingdom
- U01 AG032984/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- G-1307/PUK_/Parkinson's UK/United Kingdom
- K01 AG046374/AG/NIA NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- MR/J004758/1/MRC_/Medical Research Council/United Kingdom
- RF1 AG015473/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous


