Neuropilin-1 is upregulated in the adaptive response of prostate tumors to androgen-targeted therapies and is prognostic of metastatic progression and patient mortality
- PMID: 28092670
- PMCID: PMC5485179
- DOI: 10.1038/onc.2016.482
Neuropilin-1 is upregulated in the adaptive response of prostate tumors to androgen-targeted therapies and is prognostic of metastatic progression and patient mortality
Abstract
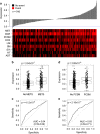
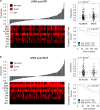
Recent evidence has implicated the transmembrane co-receptor neuropilin-1 (NRP1) in cancer progression. Primarily known as a regulator of neuronal guidance and angiogenesis, NRP1 is also expressed in multiple human malignancies, where it promotes tumor angiogenesis. However, non-angiogenic roles of NRP1 in tumor progression remain poorly characterized. In this study, we define NRP1 as an androgen-repressed gene whose expression is elevated during the adaptation of prostate tumors to androgen-targeted therapies (ATTs), and subsequent progression to metastatic castration-resistant prostate cancer (mCRPC). Using short hairpin RNA (shRNA)-mediated suppression of NRP1, we demonstrate that NRP1 regulates the mesenchymal phenotype of mCRPC cell models and the invasive and metastatic dissemination of tumor cells in vivo. In patients, immunohistochemical staining of tissue microarrays and mRNA expression analyses revealed a positive association between NRP1 expression and increasing Gleason grade, pathological T score, positive lymph node status and primary therapy failure. Furthermore, multivariate analysis of several large clinical prostate cancer (PCa) cohorts identified NRP1 expression at radical prostatectomy as an independent prognostic biomarker of biochemical recurrence after radiation therapy, metastasis and cancer-specific mortality. This study identifies NRP1 for the first time as a novel androgen-suppressed gene upregulated during the adaptive response of prostate tumors to ATTs and a prognostic biomarker of clinical metastasis and lethal PCa.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Neuropilin-1/PKC axis promotes neuroendocrine differentiation and drug resistance of prostate cancer.Br J Cancer. 2023 Mar;128(5):918-927. doi: 10.1038/s41416-022-02114-9. Epub 2022 Dec 22. Br J Cancer. 2023. PMID: 36550208 Free PMC article.
-
Differential Risk of Castration Resistance After Initial Radical Prostatectomy or Radiotherapy for Prostate Cancer.Anticancer Res. 2017 Oct;37(10):5631-5637. doi: 10.21873/anticanres.11998. Anticancer Res. 2017. PMID: 28982880
-
Neuropilin-1 promotes human glioma progression through potentiating the activity of the HGF/SF autocrine pathway.Oncogene. 2007 Aug 16;26(38):5577-86. doi: 10.1038/sj.onc.1210348. Epub 2007 Mar 19. Oncogene. 2007. PMID: 17369861 Free PMC article.
-
Cancer stem cell and mesenchymal cell cooperative actions in metastasis progression and hormone resistance in prostate cancer: Potential role of androgen and gonadotropin‑releasing hormone receptors (Review).Int J Oncol. 2020 May;56(5):1075-1082. doi: 10.3892/ijo.2020.5008. Epub 2020 Mar 5. Int J Oncol. 2020. PMID: 32319606 Review.
-
Cell death under epithelial-mesenchymal transition control in prostate cancer therapeutic response.Int J Urol. 2018 Apr;25(4):318-326. doi: 10.1111/iju.13505. Epub 2018 Jan 17. Int J Urol. 2018. PMID: 29345000 Review.
Cited by
-
Emerging Perspectives in Zinc Transporter Research in Prostate Cancer: An Updated Review.Nutrients. 2024 Jun 26;16(13):2026. doi: 10.3390/nu16132026. Nutrients. 2024. PMID: 38999774 Free PMC article. Review.
-
Differential modulation of cell morphology, migration, and Neuropilin-1 expression in cancer and non-cancer cell lines by substrate stiffness.Front Cell Dev Biol. 2024 Jun 5;12:1352233. doi: 10.3389/fcell.2024.1352233. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38903533 Free PMC article.
-
Integrative bioinformatics approach yields a novel gene expression risk model for prognosis and progression prediction in prostate cancer.J Cell Mol Med. 2024 Jun;28(11):e18405. doi: 10.1111/jcmm.18405. J Cell Mol Med. 2024. PMID: 38842134 Free PMC article.
-
Investigating the Role of SNAI1 and ZEB1 Expression in Prostate Cancer Progression and Immune Modulation of the Tumor Microenvironment.Cancers (Basel). 2024 Apr 12;16(8):1480. doi: 10.3390/cancers16081480. Cancers (Basel). 2024. PMID: 38672562 Free PMC article.
-
sBioSITe enables sensitive identification of the cell surface proteome through direct enrichment of biotinylated peptides.Clin Proteomics. 2023 Dec 5;20(1):56. doi: 10.1186/s12014-023-09445-6. Clin Proteomics. 2023. PMID: 38053024 Free PMC article.
References
-
- Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T et al. EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol 2014; 65: 467–479. - PubMed
-
- de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet 2010; 376: 1147–1154. - PubMed
-
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004; 351: 1502–1512. - PubMed
-
- James ND, Sydes MR, Clarke NW, Mason MD, Dearnaley DP, Spears MR et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 2016; 387: 1163–1177. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous


