A diverse intrinsic antibiotic resistome from a cave bacterium
- PMID: 27929110
- PMCID: PMC5155152
- DOI: 10.1038/ncomms13803
A diverse intrinsic antibiotic resistome from a cave bacterium
Abstract
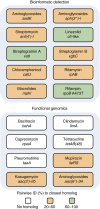
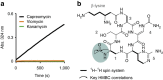
Antibiotic resistance is ancient and widespread in environmental bacteria. These are therefore reservoirs of resistance elements and reflective of the natural history of antibiotics and resistance. In a previous study, we discovered that multi-drug resistance is common in bacteria isolated from Lechuguilla Cave, an underground ecosystem that has been isolated from the surface for over 4 Myr. Here we use whole-genome sequencing, functional genomics and biochemical assays to reveal the intrinsic resistome of Paenibacillus sp. LC231, a cave bacterial isolate that is resistant to most clinically used antibiotics. We systematically link resistance phenotype to genotype and in doing so, identify 18 chromosomal resistance elements, including five determinants without characterized homologues and three mechanisms not previously shown to be involved in antibiotic resistance. A resistome comparison across related surface Paenibacillus affirms the conservation of resistance over millions of years and establishes the longevity of these genes in this genus.
Figures



Similar articles
-
The complex resistomes of Paenibacillaceae reflect diverse antibiotic chemical ecologies.ISME J. 2018 Mar;12(3):885-897. doi: 10.1038/s41396-017-0017-5. Epub 2017 Dec 19. ISME J. 2018. PMID: 29259290 Free PMC article.
-
Genomic characterization and assessment of the virulence and antibiotic resistance of the novel species Paenibacillus sp. strain VT-400, a potentially pathogenic bacterium in the oral cavity of patients with hematological malignancies.Gut Pathog. 2016 Feb 19;8:6. doi: 10.1186/s13099-016-0089-1. eCollection 2016. Gut Pathog. 2016. PMID: 26900405 Free PMC article.
-
Insight into the resistome and quorum sensing system of a divergent Acinetobacter pittii isolate from an untouched site of the Lechuguilla Cave.Access Microbiol. 2020 Jan 20;2(2):acmi000089. doi: 10.1099/acmi.0.000089. eCollection 2020. Access Microbiol. 2020. PMID: 34568753 Free PMC article.
-
Expanding the soil antibiotic resistome: exploring environmental diversity.Curr Opin Microbiol. 2007 Oct;10(5):481-9. doi: 10.1016/j.mib.2007.08.009. Epub 2007 Oct 22. Curr Opin Microbiol. 2007. PMID: 17951101 Review.
-
Antibiotic resistance in the environment: a link to the clinic?Curr Opin Microbiol. 2010 Oct;13(5):589-94. doi: 10.1016/j.mib.2010.08.005. Epub 2010 Sep 16. Curr Opin Microbiol. 2010. PMID: 20850375 Review.
Cited by
-
Revealing antibiotic resistance's ancient roots: insights from pristine ecosystems.Front Microbiol. 2024 Oct 9;15:1445155. doi: 10.3389/fmicb.2024.1445155. eCollection 2024. Front Microbiol. 2024. PMID: 39450285 Free PMC article. Review.
-
Seasonal stability of the rumen microbiome contributes to the adaptation patterns to extreme environmental conditions in grazing yak and cattle.BMC Biol. 2024 Oct 23;22(1):240. doi: 10.1186/s12915-024-02035-4. BMC Biol. 2024. PMID: 39443951 Free PMC article.
-
Inferring diet, disease and antibiotic resistance from ancient human oral microbiomes.Microb Genom. 2024 May;10(5):001251. doi: 10.1099/mgen.0.001251. Microb Genom. 2024. PMID: 38739117 Free PMC article. Review.
-
The geomicrobiology of limestone, sulfuric acid speleogenetic, and volcanic caves: basic concepts and future perspectives.Front Microbiol. 2024 Mar 20;15:1370520. doi: 10.3389/fmicb.2024.1370520. eCollection 2024. Front Microbiol. 2024. PMID: 38572233 Free PMC article. Review.
-
Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region.Antibiotics (Basel). 2023 Dec 4;12(12):1697. doi: 10.3390/antibiotics12121697. Antibiotics (Basel). 2023. PMID: 38136731 Free PMC article.
References
-
- Perry J. A., Westman E. L. & Wright G. D. The antibiotic resistome: what's new? Curr. Opin. Microbiol. 21, 45–50 (2014). - PubMed
-
- D'Costa V. M. et al.. Antibiotic resistance is ancient. Nature 477, 457–461 (2011). - PubMed
-
- D'Costa V. M., McGrann K. M., Hughes D. W. & Wright G. D. Sampling the antibiotic resistome. Science 311, 374–377 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


