A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data
- PMID: 27738200
- PMCID: PMC6078412
- DOI: 10.3852/16-042
A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data
Abstract
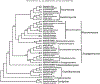
Zygomycete fungi were classified as a single phylum, Zygomycota, based on sexual reproduction by zygospores, frequent asexual reproduction by sporangia, absence of multicellular sporocarps, and production of coenocytic hyphae, all with some exceptions. Molecular phylogenies based on one or a few genes did not support the monophyly of the phylum, however, and the phylum was subsequently abandoned. Here we present phylogenetic analyses of a genome-scale data set for 46 taxa, including 25 zygomycetes and 192 proteins, and we demonstrate that zygomycetes comprise two major clades that form a paraphyletic grade. A formal phylogenetic classification is proposed herein and includes two phyla, six subphyla, four classes and 16 orders. On the basis of these results, the phyla Mucoromycota and Zoopagomycota are circumscribed. Zoopagomycota comprises Entomophtoromycotina, Kickxellomycotina and Zoopagomycotina; it constitutes the earliest diverging lineage of zygomycetes and contains species that are primarily parasites and pathogens of small animals (e.g. amoeba, insects, etc.) and other fungi, i.e. mycoparasites. Mucoromycota comprises Glomeromycotina, Mortierellomycotina, and Mucoromycotina and is sister to Dikarya. It is the more derived clade of zygomycetes and mainly consists of mycorrhizal fungi, root endophytes, and decomposers of plant material. Evolution of trophic modes, morphology, and analysis of genome-scale data are discussed.
Keywords: Entomophthoromycotina; Glomeromycotina; Kickellomycotina; Mortierellomycotina; Mucoromycota; Mucoromycotina; Zoopagomycota Zoopagomycotina; fungi; paraphyly; systematics.
© 2016 by The Mycological Society of America.
Figures

Similar articles
-
Divergent Evolution of Early Terrestrial Fungi Reveals the Evolution of Mucormycosis Pathogenicity Factors.Genome Biol Evol. 2023 Apr 6;15(4):evad046. doi: 10.1093/gbe/evad046. Genome Biol Evol. 2023. PMID: 36930540 Free PMC article.
-
A higher-level phylogenetic classification of the Fungi.Mycol Res. 2007 May;111(Pt 5):509-47. doi: 10.1016/j.mycres.2007.03.004. Epub 2007 Mar 13. Mycol Res. 2007. PMID: 17572334 Review.
-
The Fungal Tree of Life: from Molecular Systematics to Genome-Scale Phylogenies.Microbiol Spectr. 2017 Sep;5(5). doi: 10.1128/microbiolspec.FUNK-0053-2016. Microbiol Spectr. 2017. PMID: 28917057 Review.
-
Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes.BMC Evol Biol. 2006 Sep 29;6:74. doi: 10.1186/1471-2148-6-74. BMC Evol Biol. 2006. PMID: 17010206 Free PMC article.
-
Investigating Endobacteria that Thrive Within Mucoromycota.Methods Mol Biol. 2023;2605:293-323. doi: 10.1007/978-1-0716-2871-3_15. Methods Mol Biol. 2023. PMID: 36520400
Cited by
-
Flue-cured tobacco intercropping with insectary floral plants improves rhizosphere soil microbial communities and chemical properties of flue-cured tobacco.BMC Microbiol. 2024 Nov 4;24(1):446. doi: 10.1186/s12866-024-03597-7. BMC Microbiol. 2024. PMID: 39497066 Free PMC article.
-
Comparative mitochondrial genomics of Thelebolaceae in Antarctica: insights into their extremophilic adaptations and evolutionary dynamics.IMA Fungus. 2024 Oct 30;15(1):33. doi: 10.1186/s43008-024-00164-7. IMA Fungus. 2024. PMID: 39478621 Free PMC article.
-
Alleviation of NaCl Stress on Growth and Biochemical Traits of Cenchrus ciliaris L. via Arbuscular Mycorrhizal Fungi Symbiosis.Life (Basel). 2024 Oct 8;14(10):1276. doi: 10.3390/life14101276. Life (Basel). 2024. PMID: 39459576 Free PMC article.
-
Unveiling species diversity within early-diverging fungi from China I: three new species of Backusella (Backusellaceae, Mucoromycota).MycoKeys. 2024 Oct 14;109:285-304. doi: 10.3897/mycokeys.109.126029. eCollection 2024. MycoKeys. 2024. PMID: 39439597 Free PMC article.
-
Invasive Fungal Infections in Immunocompromised Conditions: Emphasis on COVID-19.Curr Microbiol. 2024 Oct 9;81(11):400. doi: 10.1007/s00284-024-03916-1. Curr Microbiol. 2024. PMID: 39384659 Review.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science 287:2185–2195, doi:10.1126/science.287.5461.2185 - DOI - PubMed
-
- Bauer R, Humber RA, Voigt K. 2014. Zygomycetous fungi: phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In: McLaughlin DJ, Spatafora JW, eds. Systematics and evolution. Part A New York: Springer-Verlag. The Mycota; VII:209–250.
-
- Benny GL, Humber RA, Voigt K. 2014. Zygomycetous fungi: phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In: McLaughlin DJ, Spatafora JW, eds. Mycota VII, part A. Systematics and evolution New York: Springer-Verlag; p 209–250.
-
- Benny GL, O’Donnell K 2000. Amoebidium parasiticum is a protozoan, not a Trichomycete. Mycologia 92:1133–1137, doi:10.2307/3761480 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

