SARS and MERS: recent insights into emerging coronaviruses
- PMID: 27344959
- PMCID: PMC7097822
- DOI: 10.1038/nrmicro.2016.81
SARS and MERS: recent insights into emerging coronaviruses
Abstract
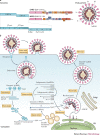
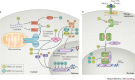
The emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 marked the second introduction of a highly pathogenic coronavirus into the human population in the twenty-first century. The continuing introductions of MERS-CoV from dromedary camels, the subsequent travel-related viral spread, the unprecedented nosocomial outbreaks and the high case-fatality rates highlight the need for prophylactic and therapeutic measures. Scientific advancements since the 2002-2003 severe acute respiratory syndrome coronavirus (SARS-CoV) pandemic allowed for rapid progress in our understanding of the epidemiology and pathogenesis of MERS-CoV and the development of therapeutics. In this Review, we detail our present understanding of the transmission and pathogenesis of SARS-CoV and MERS-CoV, and discuss the current state of development of measures to combat emerging coronaviruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Development of animal models against emerging coronaviruses: From SARS to MERS coronavirus.Virology. 2015 May;479-480:247-58. doi: 10.1016/j.virol.2015.02.030. Epub 2015 Mar 16. Virology. 2015. PMID: 25791336 Free PMC article. Review.
-
Emerging coronaviruses: first SARS, second MERS and third SARS-CoV-2: epidemiological updates of COVID-19.Infez Med. 2020 Jun 1;28(suppl 1):6-17. Infez Med. 2020. PMID: 32532933 Review.
-
Coronaviruses: severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus in travelers.Curr Opin Infect Dis. 2014 Oct;27(5):411-7. doi: 10.1097/QCO.0000000000000089. Curr Opin Infect Dis. 2014. PMID: 25033169 Review.
-
Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV.J Med Virol. 2020 May;92(5):491-494. doi: 10.1002/jmv.25709. Epub 2020 Feb 21. J Med Virol. 2020. PMID: 32056249 Free PMC article. Review.
-
A Comprehensive Review of Animal Models for Coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV.Virol Sin. 2020 Jun;35(3):290-304. doi: 10.1007/s12250-020-00252-z. Epub 2020 Jun 30. Virol Sin. 2020. PMID: 32607866 Free PMC article. Review.
Cited by
-
ACE2-independent sarbecovirus cell entry can be supported by TMPRSS2-related enzymes and can reduce sensitivity to antibody-mediated neutralization.PLoS Pathog. 2024 Nov 13;20(11):e1012653. doi: 10.1371/journal.ppat.1012653. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39536058 Free PMC article.
-
Antiseptics: An expeditious third force in the prevention and management of coronavirus diseases.Curr Res Microb Sci. 2024 Oct 16;7:100293. doi: 10.1016/j.crmicr.2024.100293. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39497935 Free PMC article. Review.
-
Pakistan's first MERS-CoV case: A wake up call for global health authorities.New Microbes New Infect. 2024 Oct 19;62:101516. doi: 10.1016/j.nmni.2024.101516. eCollection 2024 Dec. New Microbes New Infect. 2024. PMID: 39497913 Free PMC article. No abstract available.
-
Study of Otorhinolaryngological Manifestations in Symptomatic COVID-19-Positive Patients at Tertiary Health Care Hospital: A Cross-sectional Study.Int Arch Otorhinolaryngol. 2024 Jul 5;28(4):e597-e602. doi: 10.1055/s-0044-1786831. eCollection 2024 Oct. Int Arch Otorhinolaryngol. 2024. PMID: 39464359 Free PMC article.
-
Opportunity for severe and critical COVID-19 pneumonia treatment with corticosteroids: a retrospective cohort study.J Thorac Dis. 2024 Sep 30;16(9):5688-5697. doi: 10.21037/jtd-24-329. Epub 2024 Sep 11. J Thorac Dis. 2024. PMID: 39444892 Free PMC article.
References
-
- Lee N, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. - PubMed
-
- Drosten C, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous


