A paneukaryotic genomic analysis of the small GTPase RABL2 underscores the significance of recurrent gene loss in eukaryote evolution
- PMID: 26832778
- PMCID: PMC4736243
- DOI: 10.1186/s13062-016-0107-8
A paneukaryotic genomic analysis of the small GTPase RABL2 underscores the significance of recurrent gene loss in eukaryote evolution
Abstract
Background: The cilium (flagellum) is a complex cellular structure inherited from the last eukaryotic common ancestor (LECA). A large number of ciliary proteins have been characterized in a few model organisms, but their evolutionary history often remains unexplored. One such protein is the small GTPase RABL2, recently implicated in the assembly of the sperm tail in mammals.
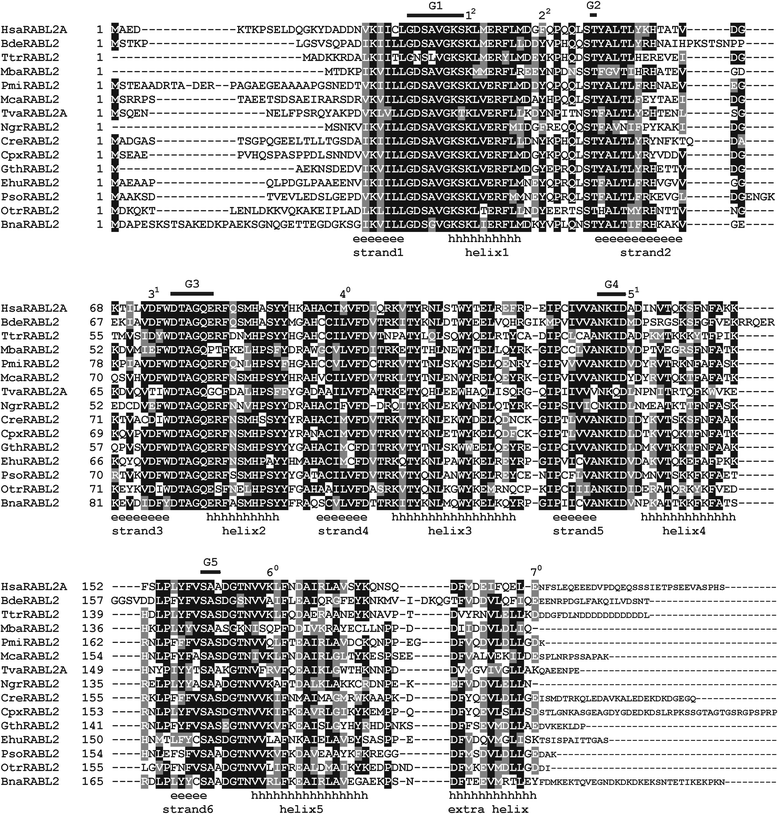
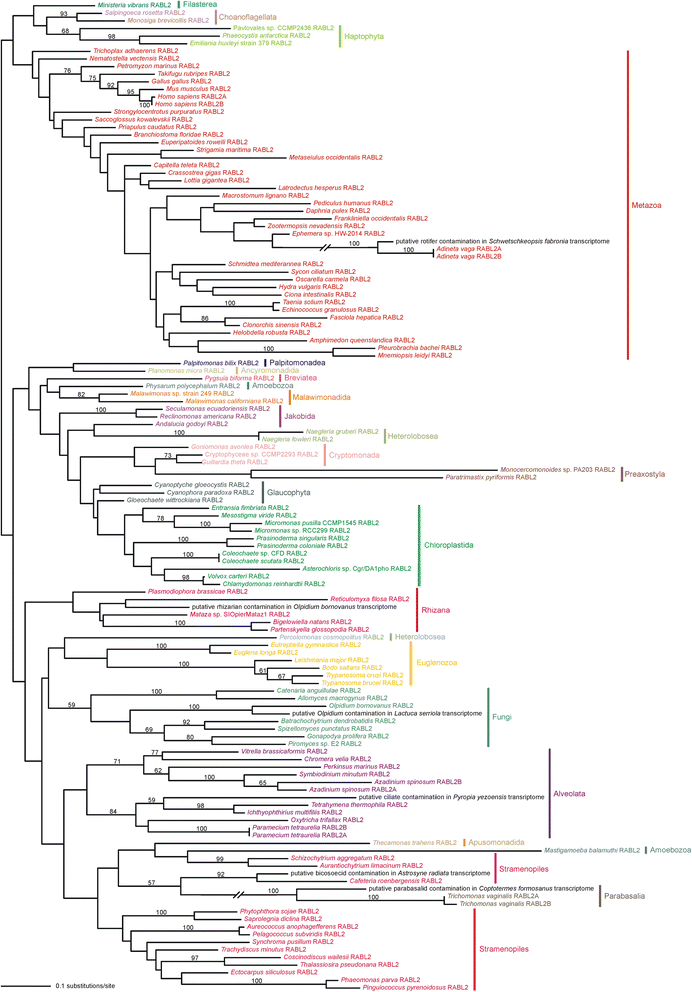
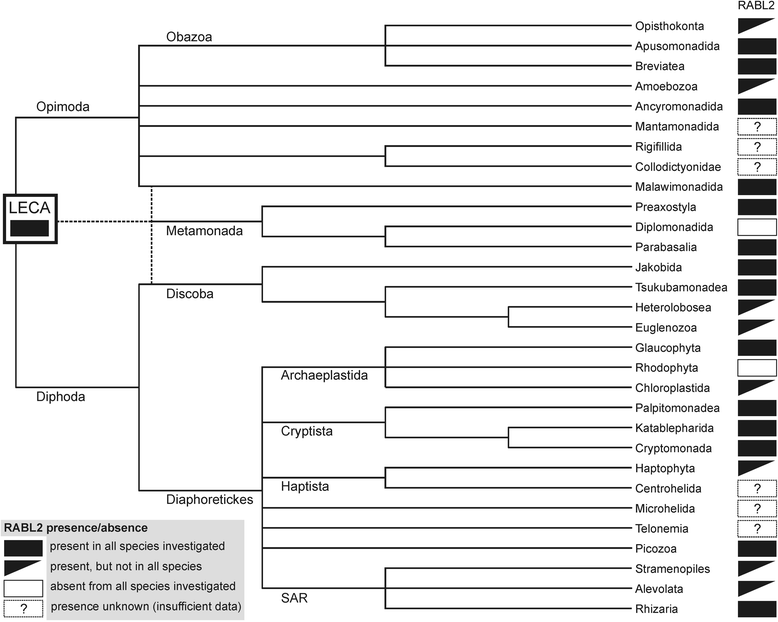
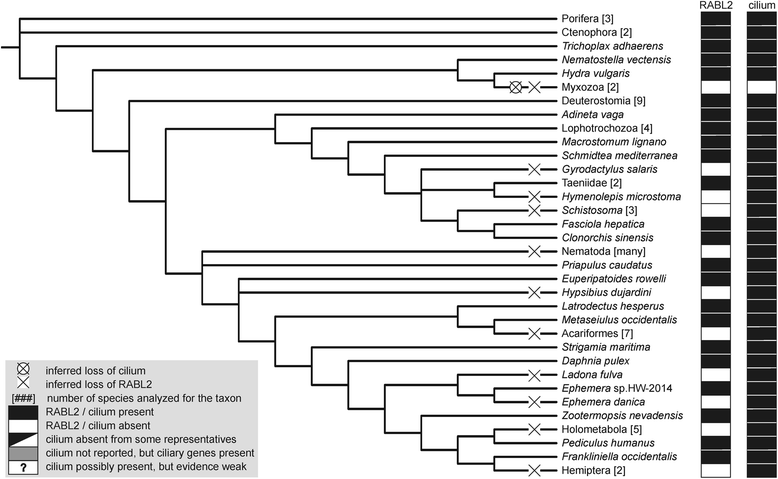
Results: Using the wealth of currently available genome and transcriptome sequences, including data from our on-going sequencing projects, we systematically analyzed the phylogenetic distribution and evolutionary history of RABL2 orthologs. Our dense taxonomic sampling revealed the presence of RABL2 genes in nearly all major eukaryotic lineages, including small "obscure" taxa such as breviates, ancyromonads, malawimonads, jakobids, picozoans, or palpitomonads. The phyletic pattern of RABL2 genes indicates that it was present already in the LECA. However, some organisms lack RABL2 as a result of secondary loss and our present sampling predicts well over 30 such independent events during the eukaryote evolution. The distribution of RABL2 genes correlates with the presence/absence of cilia: not a single well-established cilium-lacking species has retained a RABL2 ortholog. However, several ciliated taxa, most notably nematodes, some arthropods and platyhelminths, diplomonads, and ciliated subgroups of apicomplexans and embryophytes, lack RABL2 as well, suggesting some simplification in their cilium-associated functions. On the other hand, several algae currently unknown to form cilia, e.g., the "prasinophytes" of the genus Prasinoderma or the ochrophytes Pelagococcus subviridis and Pinguiococcus pyrenoidosus, turned out to encode not only RABL2, but also homologs of some hallmark ciliary proteins, suggesting the existence of a cryptic flagellated stage in their life cycles. We additionally obtained insights into the evolution of the RABL2 gene architecture, which seems to have ancestrally consisted of eight exons subsequently modified not only by lineage-specific intron loss and gain, but also by recurrent loss of the terminal exon encoding a poorly conserved C-terminal extension.
Conclusions: Our comparative analysis supports the notion that RABL2 is an ancestral component of the eukaryotic cilium and underscores the still underappreciated magnitude of recurrent gene loss, or reductive evolution in general, in the history of eukaryotic genomes and cells.
Figures

Similar articles
-
Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era.BMC Evol Biol. 2017 Sep 4;17(1):211. doi: 10.1186/s12862-017-1056-2. BMC Evol Biol. 2017. PMID: 28870165 Free PMC article.
-
Taxon-specific expansion and loss of tektins inform metazoan ciliary diversity.BMC Evol Biol. 2019 Jan 31;19(1):40. doi: 10.1186/s12862-019-1360-0. BMC Evol Biol. 2019. PMID: 30704394 Free PMC article.
-
RABL2 interacts with the intraflagellar transport-B complex and CEP19 and participates in ciliary assembly.Mol Biol Cell. 2017 Jun 15;28(12):1652-1666. doi: 10.1091/mbc.E17-01-0017. Epub 2017 Apr 20. Mol Biol Cell. 2017. PMID: 28428259 Free PMC article.
-
Concepts of the last eukaryotic common ancestor.Nat Ecol Evol. 2019 Mar;3(3):338-344. doi: 10.1038/s41559-019-0796-3. Epub 2019 Feb 18. Nat Ecol Evol. 2019. PMID: 30778187 Review.
-
Whence genes in pieces: reconstruction of the exon-intron gene structures of the last eukaryotic common ancestor and other ancestral eukaryotes.Wiley Interdiscip Rev RNA. 2013 Jan-Feb;4(1):93-105. doi: 10.1002/wrna.1143. Epub 2012 Nov 8. Wiley Interdiscip Rev RNA. 2013. PMID: 23139082 Review.
Cited by
-
Uncoupled evolution of the Polycomb system and deep origin of non-canonical PRC1.Commun Biol. 2023 Nov 10;6(1):1144. doi: 10.1038/s42003-023-05501-x. Commun Biol. 2023. PMID: 37949928 Free PMC article.
-
Phylotranscriptomics unveil a Paleoproterozoic-Mesoproterozoic origin and deep relationships of the Viridiplantae.Nat Commun. 2023 Sep 11;14(1):5542. doi: 10.1038/s41467-023-41137-5. Nat Commun. 2023. PMID: 37696791 Free PMC article.
-
The IFT81-IFT74 complex acts as an unconventional RabL2 GTPase-activating protein during intraflagellar transport.EMBO J. 2023 Sep 18;42(18):e111807. doi: 10.15252/embj.2022111807. Epub 2023 Aug 22. EMBO J. 2023. PMID: 37606072 Free PMC article.
-
Phylogenetic profiling and cellular analyses of ARL16 reveal roles in traffic of IFT140 and INPP5E.Mol Biol Cell. 2022 Apr 1;33(4):ar33. doi: 10.1091/mbc.E21-10-0509-T. Epub 2022 Feb 23. Mol Biol Cell. 2022. PMID: 35196065 Free PMC article.
-
Horizontally transferred genes in the ctenophore Mnemiopsis leidyi.PeerJ. 2018 Jun 15;6:e5067. doi: 10.7717/peerj.5067. eCollection 2018. PeerJ. 2018. PMID: 29922518 Free PMC article.
References
-
- Schmid F, Christensen ST, Pedersen LB. Cilia and Flagella. Encyclopedia of Cell Biology. 2016;2:660–76. doi: 10.1016/B978-0-12-394447-4.20064-3. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


