Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup
- PMID: 26754573
- PMCID: PMC4709632
- DOI: 10.1038/srep19157
Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup
Abstract
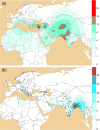
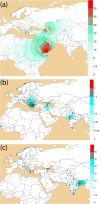
The global distribution of J2-M172 sub-haplogroups has been associated with Neolithic demic diffusion. Two branches of J2-M172, J2a-M410 and J2b-M102 make a considerable part of Y chromosome gene pool of the Indian subcontinent. We investigated the Neolithic contribution of demic dispersal from West to Indian paternal lineages, which majorly consists of haplogroups of Late Pleistocene ancestry. To accomplish this, we have analysed 3023 Y-chromosomes from different ethnic populations, of which 355 belonged to J2-M172. Comparison of our data with worldwide data, including Y-STRs of 1157 individuals and haplogroup frequencies of 6966 individuals, suggested a complex scenario that cannot be explained by a single wave of agricultural expansion from Near East to South Asia. Contrary to the widely accepted elite dominance model, we found a substantial presence of J2a-M410 and J2b-M102 haplogroups in both caste and tribal populations of India. Unlike demic spread in Eurasia, our results advocate a unique, complex and ancient arrival of J2a-M410 and J2b-M102 haplogroups into Indian subcontinent.
Figures

Similar articles
-
A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios.Proc Natl Acad Sci U S A. 2006 Jan 24;103(4):843-8. doi: 10.1073/pnas.0507714103. Epub 2006 Jan 13. Proc Natl Acad Sci U S A. 2006. PMID: 16415161 Free PMC article.
-
The Eastern side of the Westernmost Europeans: Insights from subclades within Y-chromosome haplogroup J-M304.Am J Hum Biol. 2018 Mar;30(2). doi: 10.1002/ajhb.23082. Epub 2017 Nov 29. Am J Hum Biol. 2018. PMID: 29193490
-
Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA.BMC Genet. 2006 Aug 7;7:42. doi: 10.1186/1471-2156-7-42. BMC Genet. 2006. PMID: 16893451 Free PMC article.
-
Phylogeography of mitochondrial DNA and Y-chromosome haplogroups reveal asymmetric gene flow in populations of Eastern India.Am J Phys Anthropol. 2006 Sep;131(1):84-97. doi: 10.1002/ajpa.20399. Am J Phys Anthropol. 2006. PMID: 16485297
-
Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-neolithic hunter-gatherers, Austronesian farmers, and Indian traders.Hum Biol. 2005 Feb;77(1):93-114. doi: 10.1353/hub.2005.0030. Hum Biol. 2005. PMID: 16114819
Cited by
-
Genomic Insights into the Population History of the Resande or Swedish Travelers.Genome Biol Evol. 2023 Feb 3;15(2):evad006. doi: 10.1093/gbe/evad006. Genome Biol Evol. 2023. PMID: 36655389 Free PMC article.
-
Population genetic study of 17 Y-STR Loci of the Sorani Kurds in the Province of Sulaymaniyah, Iraq.BMC Genomics. 2022 Nov 21;23(1):763. doi: 10.1186/s12864-022-09005-6. BMC Genomics. 2022. PMID: 36414939 Free PMC article.
-
Ancient Components and Recent Expansion in the Eurasian Heartland: Insights into the Revised Phylogeny of Y-Chromosomes from Central Asia.Genes (Basel). 2022 Oct 1;13(10):1776. doi: 10.3390/genes13101776. Genes (Basel). 2022. PMID: 36292661 Free PMC article.
-
Contrasting maternal and paternal genetic histories among five ethnic groups from Khyber Pakhtunkhwa, Pakistan.Sci Rep. 2022 Jan 19;12(1):1027. doi: 10.1038/s41598-022-05076-3. Sci Rep. 2022. PMID: 35046511 Free PMC article.
-
Comparative Y-chromosome analysis among Cypriots in the context of historical events and migrations.PLoS One. 2021 Aug 23;16(8):e0255140. doi: 10.1371/journal.pone.0255140. eCollection 2021. PLoS One. 2021. PMID: 34424929 Free PMC article.
References
-
- Misra V. N. Prehistoric human colonisation of India. J. Biosci. 26, 491–531 (2001). - PubMed
-
- Kivisild T. et al. In Genomic Diversity: Applications in Human Population Genetics (eds Papiha S. S., Deka R. & Chakraborty R.) Ch. 11, 135–152 (Kluwer, New York, 1999).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources


