Foxd3 Promotes Exit from Naive Pluripotency through Enhancer Decommissioning and Inhibits Germline Specification
- PMID: 26748758
- PMCID: PMC5048917
- DOI: 10.1016/j.stem.2015.09.010
Foxd3 Promotes Exit from Naive Pluripotency through Enhancer Decommissioning and Inhibits Germline Specification
Abstract
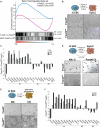
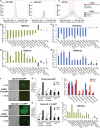
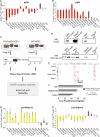
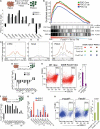
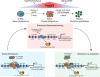
Following implantation, mouse epiblast cells transit from a naive to a primed state in which they are competent for both somatic and primordial germ cell (PGC) specification. Using mouse embryonic stem cells as an in vitro model to study the transcriptional regulatory principles orchestrating peri-implantation development, here we show that the transcription factor Foxd3 is necessary for exit from naive pluripotency and progression to a primed pluripotent state. During this transition, Foxd3 acts as a repressor that dismantles a significant fraction of the naive pluripotency expression program through decommissioning of active enhancers associated with key naive pluripotency and early germline genes. Subsequently, Foxd3 needs to be silenced in primed pluripotent cells to allow re-activation of relevant genes required for proper PGC specification. Our findings therefore uncover a cycle of activation and deactivation of Foxd3 required for exit from naive pluripotency and subsequent PGC specification.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
The paradox of Foxd3: how does it function in pluripotency and differentiation of embryonic stem cells?Stem Cell Investig. 2016 Nov 4;3:73. doi: 10.21037/sci.2016.09.20. eCollection 2016. Stem Cell Investig. 2016. PMID: 27868055 Free PMC article.
Similar articles
-
Stepwise differentiation from naïve state pluripotent stem cells to functional primordial germ cells through an epiblast-like state.Methods Mol Biol. 2013;1074:175-83. doi: 10.1007/978-1-62703-628-3_13. Methods Mol Biol. 2013. PMID: 23975813
-
FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity.Cell Stem Cell. 2016 Jan 7;18(1):104-17. doi: 10.1016/j.stem.2015.10.003. Cell Stem Cell. 2016. PMID: 26748757 Free PMC article.
-
NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers.Nature. 2016 Jan 21;529(7586):403-407. doi: 10.1038/nature16480. Epub 2016 Jan 11. Nature. 2016. PMID: 26751055 Free PMC article.
-
Mapping the route from naive pluripotency to lineage specification.Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130540. doi: 10.1098/rstb.2013.0540. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25349449 Free PMC article. Review.
-
Germ cell specification in mice.Science. 2007 Apr 20;316(5823):394-6. doi: 10.1126/science.1137545. Science. 2007. PMID: 17446386 Review.
Cited by
-
Poised PABP-RNA hubs implement signal-dependent mRNA decay in development.Nat Struct Mol Biol. 2024 Sep;31(9):1439-1447. doi: 10.1038/s41594-024-01363-x. Epub 2024 Jul 25. Nat Struct Mol Biol. 2024. PMID: 39054355 Free PMC article.
-
BET activity plays an essential role in control of stem cell attributes in Xenopus.Development. 2024 Jul 1;151(13):dev202990. doi: 10.1242/dev.202990. Epub 2024 Jul 3. Development. 2024. PMID: 38884356 Free PMC article.
-
Regulation of Myc transcription by an enhancer cluster dedicated to pluripotency and early embryonic expression.Nat Commun. 2024 May 10;15(1):3931. doi: 10.1038/s41467-024-48258-5. Nat Commun. 2024. PMID: 38729993 Free PMC article.
-
Cellular reprogramming in vivo initiated by SOX4 pioneer factor activity.Nat Commun. 2024 Feb 26;15(1):1761. doi: 10.1038/s41467-024-45939-z. Nat Commun. 2024. PMID: 38409161 Free PMC article.
-
Transcriptional repression upon S phase entry protects genome integrity in pluripotent cells.Nat Struct Mol Biol. 2023 Oct;30(10):1561-1570. doi: 10.1038/s41594-023-01092-7. Epub 2023 Sep 11. Nat Struct Mol Biol. 2023. PMID: 37696959
References
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. - PubMed
-
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. - PubMed
-
- Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 2002;277:40958–40966. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


