Accuracy of Genomic Selection in a Rice Synthetic Population Developed for Recurrent Selection Breeding
- PMID: 26313446
- PMCID: PMC4551487
- DOI: 10.1371/journal.pone.0136594
Accuracy of Genomic Selection in a Rice Synthetic Population Developed for Recurrent Selection Breeding
Erratum in
-
Correction: Accuracy of Genomic Selection in a Rice Synthetic Population Developed for Recurrent Selection Breeding.PLoS One. 2016 May 19;11(5):e0154976. doi: 10.1371/journal.pone.0154976. eCollection 2016. PLoS One. 2016. PMID: 27195492 Free PMC article.
Abstract
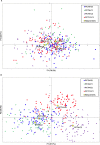
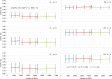
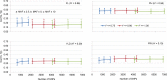
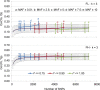
Genomic selection (GS) is a promising strategy for enhancing genetic gain. We investigated the accuracy of genomic estimated breeding values (GEBV) in four inter-related synthetic populations that underwent several cycles of recurrent selection in an upland rice-breeding program. A total of 343 S2:4 lines extracted from those populations were phenotyped for flowering time, plant height, grain yield and panicle weight, and genotyped with an average density of one marker per 44.8 kb. The relative effect of the linkage disequilibrium (LD) and minor allele frequency (MAF) thresholds for selecting markers, the relative size of the training population (TP) and of the validation population (VP), the selected trait and the genomic prediction models (frequentist and Bayesian) on the accuracy of GEBVs was investigated in 540 cross validation experiments with 100 replicates. The effect of kinship between the training and validation populations was tested in an additional set of 840 cross validation experiments with a single genomic prediction model. LD was high (average r2 = 0.59 at 25 kb) and decreased slowly, distribution of allele frequencies at individual loci was markedly skewed toward unbalanced frequencies (MAF average value 15.2% and median 9.6%), and differentiation between the four synthetic populations was low (FST ≤0.06). The accuracy of GEBV across all cross validation experiments ranged from 0.12 to 0.54 with an average of 0.30. Significant differences in accuracy were observed among the different levels of each factor investigated. Phenotypic traits had the biggest effect, and the size of the incidence matrix had the smallest. Significant first degree interaction was observed for GEBV accuracy between traits and all the other factors studied, and between prediction models and LD, MAF and composition of the TP. The potential of GS to accelerate genetic gain and breeding options to increase the accuracy of predictions are discussed.
Conflict of interest statement
Figures
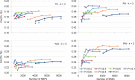
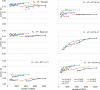
Similar articles
-
Genomic prediction models for traits differing in heritability for soybean, rice, and maize.BMC Plant Biol. 2022 Feb 26;22(1):87. doi: 10.1186/s12870-022-03479-y. BMC Plant Biol. 2022. PMID: 35219296 Free PMC article.
-
Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines.PLoS Genet. 2015 Feb 17;11(2):e1004982. doi: 10.1371/journal.pgen.1004982. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25689273 Free PMC article.
-
Rice diversity panel provides accurate genomic predictions for complex traits in the progenies of biparental crosses involving members of the panel.Theor Appl Genet. 2018 Feb;131(2):417-435. doi: 10.1007/s00122-017-3011-4. Epub 2017 Nov 14. Theor Appl Genet. 2018. PMID: 29138904 Free PMC article.
-
Invited review: Genomic selection in dairy cattle: progress and challenges.J Dairy Sci. 2009 Feb;92(2):433-43. doi: 10.3168/jds.2008-1646. J Dairy Sci. 2009. PMID: 19164653 Review.
-
Genomic Selection in Plant Breeding: Methods, Models, and Perspectives.Trends Plant Sci. 2017 Nov;22(11):961-975. doi: 10.1016/j.tplants.2017.08.011. Epub 2017 Sep 28. Trends Plant Sci. 2017. PMID: 28965742 Review.
Cited by
-
Genomic selection for tolerance to aluminum toxicity in a synthetic population of upland rice.PLoS One. 2024 Aug 22;19(8):e0307009. doi: 10.1371/journal.pone.0307009. eCollection 2024. PLoS One. 2024. PMID: 39173048 Free PMC article.
-
Effective population size in field pea.BMC Genomics. 2024 Jul 16;25(1):695. doi: 10.1186/s12864-024-10587-6. BMC Genomics. 2024. PMID: 39009980 Free PMC article.
-
Genetic gains in IRRI's rice salinity breeding and elite panel development as a future breeding resource.Theor Appl Genet. 2024 Jan 31;137(2):37. doi: 10.1007/s00122-024-04545-9. Theor Appl Genet. 2024. PMID: 38294550 Free PMC article.
-
The Era of Plant Breeding: Conventional Breeding to Genomics-assisted Breeding for Crop Improvement.Curr Genomics. 2023 Jun 23;24(1):24-35. doi: 10.2174/1389202924666230517115912. Curr Genomics. 2023. PMID: 37920729 Free PMC article. Review.
-
Optimization of Multi-Generation Multi-location Genomic Prediction Models for Recurrent Genomic Selection in an Upland Rice Population.Rice (N Y). 2023 Sep 27;16(1):43. doi: 10.1186/s12284-023-00661-0. Rice (N Y). 2023. PMID: 37758969 Free PMC article.
References
-
- Lorenz AJ, CHAO S, Asoro FG, Heffner EL, Hayashi T, Iwata H, et al. Genomic Selection in Plant Breeding: Knowledge and Prospects 1st ed. Advances in Agronomy. Elsevier Inc; 2011. pp. 77–123. 10.1016/B978-0-12-385531-2.00002-5 - DOI
-
- Bernardo R, Yu J. Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop Science. 2007;47: 1082 10.2135/cropsci2006.11.0690 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous


