Alternative Splice in Alternative Lice
- PMID: 26169943
- PMCID: PMC4576711
- DOI: 10.1093/molbev/msv151
Alternative Splice in Alternative Lice
Abstract
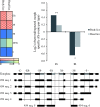
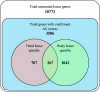
Genomic and transcriptomics analyses have revealed human head and body lice to be almost genetically identical; although con-specific, they nevertheless occupy distinct ecological niches and have differing feeding patterns. Most importantly, while head lice are not known to be vector competent, body lice can transmit three serious bacterial diseases; epidemictyphus, trench fever, and relapsing fever. In order to gain insights into the molecular bases for these differences, we analyzed alternative splicing (AS) using next-generation sequencing data for one strain of head lice and one strain of body lice. We identified a total of 3,598 AS events which were head or body lice specific. Exon skipping AS events were overrepresented among both head and body lice, whereas intron retention events were underrepresented in both. However, both the enrichment of exon skipping and the underrepresentation of intron retention are significantly stronger in body lice compared with head lice. Genes containing body louse-specific AS events were found to be significantly enriched for functions associated with development of the nervous system, salivary gland, trachea, and ovarian follicle cells, as well as regulation of transcription. In contrast, no functional categories were overrepresented among genes with head louse-specific AS events. Together, our results constitute the first evidence for transcript pool differences in head and body lice, providing insights into molecular adaptations that enabled human lice to adapt to clothing, and representing a powerful illustration of the pivotal role AS can play in functional adaptation.
Keywords: alternative splicing; body lice; head lice; human parasite; phenotype evolution; transcriptomics.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Genotyping of human lice suggests multiple emergencies of body lice from local head louse populations.PLoS Negl Trop Dis. 2010 Mar 23;4(3):e641. doi: 10.1371/journal.pntd.0000641. PLoS Negl Trop Dis. 2010. PMID: 20351779 Free PMC article.
-
What's in a name: the taxonomic status of human head and body lice.Mol Phylogenet Evol. 2008 Jun;47(3):1203-16. doi: 10.1016/j.ympev.2008.03.014. Epub 2008 Mar 16. Mol Phylogenet Evol. 2008. PMID: 18434207
-
The origin and distribution of human lice in the world.Infect Genet Evol. 2014 Apr;23:209-17. doi: 10.1016/j.meegid.2014.01.017. Epub 2014 Feb 11. Infect Genet Evol. 2014. PMID: 24524985 Review.
-
Differential gene expression in laboratory strains of human head and body lice when challenged with Bartonella quintana, a pathogenic bacterium.Insect Mol Biol. 2014 Apr;23(2):244-54. doi: 10.1111/imb.12077. Epub 2014 Jan 9. Insect Mol Biol. 2014. PMID: 24404961 Free PMC article.
-
Where Are We With Human Lice? A Review of the Current State of Knowledge.Front Cell Infect Microbiol. 2020 Jan 21;9:474. doi: 10.3389/fcimb.2019.00474. eCollection 2019. Front Cell Infect Microbiol. 2020. PMID: 32039050 Free PMC article. Review.
Cited by
-
Lack of paternal silencing and ecotype-specific expression in head and body lice hybrids.Evol Lett. 2024 Feb 6;8(3):455-465. doi: 10.1093/evlett/qrae003. eCollection 2024 Jun. Evol Lett. 2024. PMID: 38818422 Free PMC article.
-
Nucleotide-level distance metrics to quantify alternative splicing implemented in TranD.Nucleic Acids Res. 2024 Mar 21;52(5):e28. doi: 10.1093/nar/gkae056. Nucleic Acids Res. 2024. PMID: 38340337 Free PMC article.
-
Comprehensive isoform-level analysis reveals the contribution of alternative isoforms to venom evolution and repertoire diversity.Genome Res. 2023 Sep;33(9):1554-1567. doi: 10.1101/gr.277707.123. Epub 2023 Oct 5. Genome Res. 2023. PMID: 37798117 Free PMC article.
-
Alternative splicing as a source of phenotypic diversity.Nat Rev Genet. 2022 Nov;23(11):697-710. doi: 10.1038/s41576-022-00514-4. Epub 2022 Jul 12. Nat Rev Genet. 2022. PMID: 35821097 Review.
-
Human pediculosis, a global public health problem.Infect Dis Poverty. 2022 May 26;11(1):58. doi: 10.1186/s40249-022-00986-w. Infect Dis Poverty. 2022. PMID: 35619191 Free PMC article. Review.
References
-
- Ast G. 2004. How did alternative splicing evolve? Nat Rev Genet. 5:773–782. - PubMed
-
- Bacot A. 2009. A contribution to the bionomics of Pediculus humanus (Vestimenti) and Pediculus capitis . Parasitology 9:228. - PubMed
-
- Baehrecke EH. 2002. How death shapes life during development. Nat Rev Mol Cell Biol. 3:779–787. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


