The SMARCA2/4 ATPase Domain Surpasses the Bromodomain as a Drug Target in SWI/SNF-Mutant Cancers: Insights from cDNA Rescue and PFI-3 Inhibitor Studies
- PMID: 26139243
- PMCID: PMC4755107
- DOI: 10.1158/0008-5472.CAN-14-3798
The SMARCA2/4 ATPase Domain Surpasses the Bromodomain as a Drug Target in SWI/SNF-Mutant Cancers: Insights from cDNA Rescue and PFI-3 Inhibitor Studies
Abstract
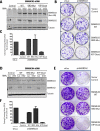
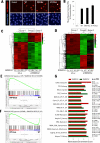
The SWI/SNF multisubunit complex modulates chromatin structure through the activity of two mutually exclusive catalytic subunits, SMARCA2 and SMARCA4, which both contain a bromodomain and an ATPase domain. Using RNAi, cancer-specific vulnerabilities have been identified in SWI/SNF-mutant tumors, including SMARCA4-deficient lung cancer; however, the contribution of conserved, druggable protein domains to this anticancer phenotype is unknown. Here, we functionally deconstruct the SMARCA2/4 paralog dependence of cancer cells using bioinformatics, genetic, and pharmacologic tools. We evaluate a selective SMARCA2/4 bromodomain inhibitor (PFI-3) and characterize its activity in chromatin-binding and cell-functional assays focusing on cells with altered SWI/SNF complex (e.g., lung, synovial sarcoma, leukemia, and rhabdoid tumors). We demonstrate that PFI-3 is a potent, cell-permeable probe capable of displacing ectopically expressed, GFP-tagged SMARCA2-bromodomain from chromatin, yet contrary to target knockdown, the inhibitor fails to display an antiproliferative phenotype. Mechanistically, the lack of pharmacologic efficacy is reconciled by the failure of bromodomain inhibition to displace endogenous, full-length SMARCA2 from chromatin as determined by in situ cell extraction, chromatin immunoprecipitation, and target gene expression studies. Furthermore, using inducible RNAi and cDNA complementation (bromodomain- and ATPase-dead constructs), we unequivocally identify the ATPase domain, and not the bromodomain of SMARCA2, as the relevant therapeutic target with the catalytic activity suppressing defined transcriptional programs. Taken together, our complementary genetic and pharmacologic studies exemplify a general strategy for multidomain protein drug-target validation and in case of SMARCA2/4 highlight the potential for drugging the more challenging helicase/ATPase domain to deliver on the promise of synthetic-lethality therapy.
©2015 American Association for Cancer Research.
Figures

Similar articles
-
Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type.J Pathol. 2016 Feb;238(3):389-400. doi: 10.1002/path.4633. Epub 2015 Dec 21. J Pathol. 2016. PMID: 26356327 Free PMC article.
-
SWI/SNF Complex-deficient Undifferentiated/Rhabdoid Carcinomas of the Gastrointestinal Tract: A Series of 13 Cases Highlighting Mutually Exclusive Loss of SMARCA4 and SMARCA2 and Frequent Co-inactivation of SMARCB1 and SMARCA2.Am J Surg Pathol. 2016 Apr;40(4):544-53. doi: 10.1097/PAS.0000000000000554. Am J Surg Pathol. 2016. PMID: 26551623
-
Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation.Mol Cell Biol. 2014 Mar;34(6):1136-44. doi: 10.1128/MCB.01372-13. Epub 2014 Jan 13. Mol Cell Biol. 2014. PMID: 24421395 Free PMC article.
-
Synthetic lethality: targeting SMARCA2 ATPase in SMARCA4-deficient tumors - a review of patent literature from 2019-30 June 2023.Expert Opin Ther Pat. 2024 Mar;34(3):159-169. doi: 10.1080/13543776.2024.2338111. Epub 2024 Apr 5. Expert Opin Ther Pat. 2024. PMID: 38578210 Review.
-
[Chromatin remodeling defects and cancer: the SWI/SNF example].Bull Cancer. 2012 Dec;99(12):1133-40. doi: 10.1684/bdc.2012.1664. Bull Cancer. 2012. PMID: 23222069 Review. French.
Cited by
-
[Research Progress on SMARCA4 Mutation Non-small Cell Lung Cancer].Zhongguo Fei Ai Za Zhi. 2024 Sep 20;27(9):704-710. doi: 10.3779/j.issn.1009-3419.2024.102.32. Zhongguo Fei Ai Za Zhi. 2024. PMID: 39492586 Free PMC article. Review. Chinese.
-
Chromatin remodellers as therapeutic targets.Nat Rev Drug Discov. 2024 Sep;23(9):661-681. doi: 10.1038/s41573-024-00978-5. Epub 2024 Jul 16. Nat Rev Drug Discov. 2024. PMID: 39014081 Review.
-
New Horizons of Synthetic Lethality in Cancer: Current Development and Future Perspectives.J Med Chem. 2024 Jul 25;67(14):11488-11521. doi: 10.1021/acs.jmedchem.4c00113. Epub 2024 Jul 2. J Med Chem. 2024. PMID: 38955347 Free PMC article. Review.
-
Unlocking cellular plasticity: enhancing human iPSC reprogramming through bromodomain inhibition and extracellular matrix gene expression regulation.Stem Cells. 2024 Aug 1;42(8):706-719. doi: 10.1093/stmcls/sxae039. Stem Cells. 2024. PMID: 38825983
-
Targeting SWI/SNF Complexes in Cancer: Pharmacological Approaches and Implications.Epigenomes. 2024 Feb 4;8(1):7. doi: 10.3390/epigenomes8010007. Epigenomes. 2024. PMID: 38390898 Free PMC article. Review.
References
-
- Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31(34):3827–44. - PubMed
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. - PubMed
-
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous


