Incorporation of spike and membrane glycoproteins into coronavirus virions
- PMID: 25855243
- PMCID: PMC4411675
- DOI: 10.3390/v7041700
Incorporation of spike and membrane glycoproteins into coronavirus virions
Abstract
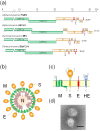
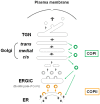
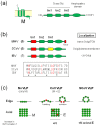
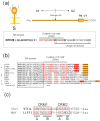
The envelopes of coronaviruses (CoVs) contain primarily three proteins; the two major glycoproteins spike (S) and membrane (M), and envelope (E), a non-glycosylated protein. Unlike other enveloped viruses, CoVs bud and assemble at the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC). For efficient virion assembly, these proteins must be targeted to the budding site and to interact with each other or the ribonucleoprotein. Thus, the efficient incorporation of viral envelope proteins into CoV virions depends on protein trafficking and protein-protein interactions near the ERGIC. The goal of this review is to summarize recent findings on the mechanism of incorporation of the M and S glycoproteins into the CoV virion, focusing on protein trafficking and protein-protein interactions.
Figures

Similar articles
-
Envelope glycoprotein interactions in coronavirus assembly.J Cell Biol. 1995 Oct;131(2):339-49. doi: 10.1083/jcb.131.2.339. J Cell Biol. 1995. PMID: 7593163 Free PMC article.
-
Vaccinia Virus Glycoproteins A33, A34, and B5 Form a Complex for Efficient Endoplasmic Reticulum to trans-Golgi Network Transport.J Virol. 2020 Mar 17;94(7):e02155-19. doi: 10.1128/JVI.02155-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31941777 Free PMC article.
-
Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E.J Gen Virol. 2005 May;86(Pt 5):1423-1434. doi: 10.1099/vir.0.80671-0. J Gen Virol. 2005. PMID: 15831954
-
The pseudotypic paradox.J Gen Virol. 1982 Nov;63 (Pt 1):15-24. doi: 10.1099/0022-1317-63-1-15. J Gen Virol. 1982. PMID: 6757385 Review.
-
[Coronaviruses].Uirusu. 2011 Dec;61(2):205-10. doi: 10.2222/jsv.61.205. Uirusu. 2011. PMID: 22916567 Review. Japanese.
Cited by
-
The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis.bioRxiv [Preprint]. 2024 Jul 16:2024.06.02.597051. doi: 10.1101/2024.06.02.597051. bioRxiv. 2024. PMID: 38895409 Free PMC article. Preprint.
-
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity.Nat Commun. 2024 May 16;15(1):4162. doi: 10.1038/s41467-024-48503-x. Nat Commun. 2024. PMID: 38755139 Free PMC article.
-
The PACS-2 protein and trafficking motifs in CCHFV Gn and Gc cytoplasmic domains govern CCHFV assembly.Emerg Microbes Infect. 2024 Dec;13(1):2348508. doi: 10.1080/22221751.2024.2348508. Epub 2024 Jun 6. Emerg Microbes Infect. 2024. PMID: 38661085 Free PMC article.
-
N-glycosylation of the SARS-CoV-2 spike protein at Asn331 and Asn343 is involved in spike-ACE2 binding, virus entry, and regulation of IL-6.Microbiol Immunol. 2024 May;68(5):165-178. doi: 10.1111/1348-0421.13121. Epub 2024 Mar 6. Microbiol Immunol. 2024. PMID: 38444370
-
A single C-terminal residue controls SARS-CoV-2 spike trafficking and incorporation into VLPs.Nat Commun. 2023 Dec 15;14(1):8358. doi: 10.1038/s41467-023-44076-3. Nat Commun. 2023. PMID: 38102143 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


