Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
- PMID: 25494202
- PMCID: PMC4420636
- DOI: 10.1038/nature14136
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
Abstract
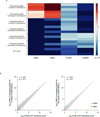
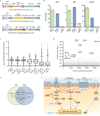
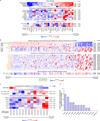
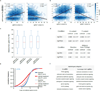
Systematic interrogation of gene function requires the ability to perturb gene expression in a robust and generalizable manner. Here we describe structure-guided engineering of a CRISPR-Cas9 complex to mediate efficient transcriptional activation at endogenous genomic loci. We used these engineered Cas9 activation complexes to investigate single-guide RNA (sgRNA) targeting rules for effective transcriptional activation, to demonstrate multiplexed activation of ten genes simultaneously, and to upregulate long intergenic non-coding RNA (lincRNA) transcripts. We also synthesized a library consisting of 70,290 guides targeting all human RefSeq coding isoforms to screen for genes that, upon activation, confer resistance to a BRAF inhibitor. The top hits included genes previously shown to be able to confer resistance, and novel candidates were validated using individual sgRNA and complementary DNA overexpression. A gene expression signature based on the top screening hits correlated with markers of BRAF inhibitor resistance in cell lines and patient-derived samples. These results collectively demonstrate the potential of Cas9-based activators as a powerful genetic perturbation technology.
Figures

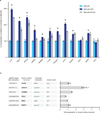
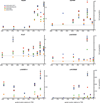
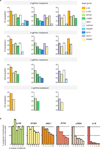
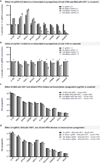


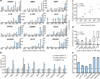
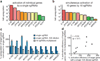


Comment in
-
Genomics: CRISPR engineering turns on genes.Nature. 2015 Jan 29;517(7536):560-2. doi: 10.1038/517560a. Nature. 2015. PMID: 25631441 No abstract available.
-
CRISPR gain-of-function screens.Nat Methods. 2015 Feb;12(2):102-3. doi: 10.1038/nmeth.3274. Nat Methods. 2015. PMID: 25798470 No abstract available.
-
Toward Whole-Transcriptome Editing with CRISPR-Cas9.Mol Cell. 2015 May 21;58(4):560-2. doi: 10.1016/j.molcel.2015.05.016. Mol Cell. 2015. PMID: 26000839
-
CRISPR-mediated multiplexed genetic manipulation.Oncotarget. 2016 Dec 6;7(49):80103-80104. doi: 10.18632/oncotarget.13371. Oncotarget. 2016. PMID: 27863416 Free PMC article. No abstract available.
-
How to switch on genes with CRISPR/Cas9?Acta Physiol (Oxf). 2018 Sep;224(1):e13087. doi: 10.1111/apha.13087. Epub 2018 May 24. Acta Physiol (Oxf). 2018. PMID: 29729207 No abstract available.
Similar articles
-
Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood.Nature. 2017 Aug 17;548(7667):343-346. doi: 10.1038/nature23451. Epub 2017 Aug 11. Nature. 2017. PMID: 28792927 Free PMC article.
-
Genome-scale CRISPR-Cas9 knockout screening in human cells.Science. 2014 Jan 3;343(6166):84-87. doi: 10.1126/science.1247005. Epub 2013 Dec 12. Science. 2014. PMID: 24336571 Free PMC article.
-
High-resolution interrogation of functional elements in the noncoding genome.Science. 2016 Sep 30;353(6307):1545-1549. doi: 10.1126/science.aaf7613. Science. 2016. PMID: 27708104 Free PMC article.
-
Cas9, Cpf1 and C2c1/2/3-What's next?Bioengineered. 2017 May 4;8(3):265-273. doi: 10.1080/21655979.2017.1282018. Epub 2017 Jan 31. Bioengineered. 2017. PMID: 28140746 Free PMC article. Review.
-
CRISPR/Cas9 system as an innovative genetic engineering tool: Enhancements in sequence specificity and delivery methods.Biochim Biophys Acta. 2015 Dec;1856(2):234-43. doi: 10.1016/j.bbcan.2015.09.003. Epub 2015 Nov 11. Biochim Biophys Acta. 2015. PMID: 26434948 Review.
Cited by
-
RLS-associated MEIS transcription factors control distinct processes in human neural stem cells.Sci Rep. 2024 Nov 22;14(1):28986. doi: 10.1038/s41598-024-80266-9. Sci Rep. 2024. PMID: 39578497 Free PMC article.
-
A benchmarked, high-efficiency prime editing platform for multiplexed dropout screening.Nat Methods. 2024 Nov 19. doi: 10.1038/s41592-024-02502-4. Online ahead of print. Nat Methods. 2024. PMID: 39562753
-
Barcoding of small extracellular vesicles with CRISPR-gRNA enables comprehensive, subpopulation-specific analysis of their biogenesis and release regulators.Nat Commun. 2024 Nov 19;15(1):9777. doi: 10.1038/s41467-024-53736-x. Nat Commun. 2024. PMID: 39562573 Free PMC article.
-
Active enhancers: recent research advances and insights into disease.Biol Direct. 2024 Nov 12;19(1):112. doi: 10.1186/s13062-024-00559-x. Biol Direct. 2024. PMID: 39533395 Free PMC article. Review.
-
TMED inhibition suppresses cell surface PD-1 expression and overcomes T cell dysfunction.J Immunother Cancer. 2024 Nov 7;12(11):e010145. doi: 10.1136/jitc-2024-010145. J Immunother Cancer. 2024. PMID: 39510795 Free PMC article.
References
-
- Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. - PubMed
-
- Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. - PubMed
-
- Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14628–14633. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials


