Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation
- PMID: 25426264
- PMCID: PMC4243145
- DOI: 10.4331/wjbc.v5.i4.398
Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation
Abstract
Pseudouridines (Ψs) are the most abundant and highly conserved modified nucleotides found in various stable RNAs of all organisms. Most Ψs are clustered in regions that are functionally important for pre-mRNA splicing. Ψ has an extra hydrogen bond donor that endows RNA molecules with distinct properties that contribute significantly to RNA-mediated cellular processes. Experimental data indicate that spliceosomal snRNA pseudouridylation can be catalyzed by both RNA-dependent and RNA-independent mechanisms. Recent work has also demonstrated that pseudouridylation can be induced at novel positions under stress conditions, suggesting a regulatory role for Ψ.
Keywords: Box H/ACA ribonucleoprotein; Induced RNA modification; Pre-mRNA splicing; Pseudouridine; U2 snRNA.
Figures
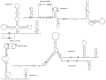

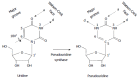
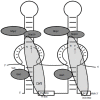
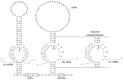
Similar articles
-
Pseudouridines in spliceosomal snRNAs.Protein Cell. 2011 Sep;2(9):712-25. doi: 10.1007/s13238-011-1087-1. Epub 2011 Oct 6. Protein Cell. 2011. PMID: 21976061 Free PMC article. Review.
-
Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):571-81. doi: 10.1002/wrna.77. Epub 2011 Feb 18. Wiley Interdiscip Rev RNA. 2011. PMID: 21957045 Free PMC article. Review.
-
An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes.RNA. 2002 Dec;8(12):1515-25. RNA. 2002. PMID: 12515384 Free PMC article.
-
Human box H/ACA pseudouridylation guide RNA machinery.Mol Cell Biol. 2004 Jul;24(13):5797-807. doi: 10.1128/MCB.24.13.5797-5807.2004. Mol Cell Biol. 2004. PMID: 15199136 Free PMC article.
-
Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo.Nucleic Acids Res. 2007;35(2):550-8. doi: 10.1093/nar/gkl1084. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17169984 Free PMC article.
Cited by
-
Circular RNAs: A New Approach to Multiple Sclerosis.Biomedicines. 2023 Oct 24;11(11):2883. doi: 10.3390/biomedicines11112883. Biomedicines. 2023. PMID: 38001884 Free PMC article. Review.
-
Transcriptome-wide analysis of pseudouridylation in Drosophila melanogaster.G3 (Bethesda). 2023 Mar 9;13(3):jkac333. doi: 10.1093/g3journal/jkac333. G3 (Bethesda). 2023. PMID: 36534986 Free PMC article.
-
The plant epitranscriptome: revisiting pseudouridine and 2'-O-methyl RNA modifications.Plant Biotechnol J. 2022 Jul;20(7):1241-1256. doi: 10.1111/pbi.13829. Epub 2022 May 11. Plant Biotechnol J. 2022. PMID: 35445501 Free PMC article. Review.
-
Regulation of RNA Splicing: Aberrant Splicing Regulation and Therapeutic Targets in Cancer.Cells. 2021 Apr 16;10(4):923. doi: 10.3390/cells10040923. Cells. 2021. PMID: 33923658 Free PMC article. Review.
-
Spliceosomal snRNA Epitranscriptomics.Front Genet. 2021 Mar 2;12:652129. doi: 10.3389/fgene.2021.652129. eCollection 2021. Front Genet. 2021. PMID: 33737950 Free PMC article. Review.
References
-
- Crick F. Split genes and RNA splicing. Science. 1979;204:264–271. - PubMed
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–137. - PubMed
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


