Structural insights into eukaryotic DNA replication
- PMID: 25202305
- PMCID: PMC4142720
- DOI: 10.3389/fmicb.2014.00444
Structural insights into eukaryotic DNA replication
Abstract
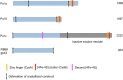
THREE DNA POLYMERASES OF THE B FAMILY FUNCTION AT THE REPLICATION FORK IN EUKARYOTIC CELLS: DNA polymerases α, δ, and ε. DNA polymerase α, an heterotetramer composed of two primase subunits and two polymerase subunits, initiates replication. DNA polymerases δ and ε elongate the primers generated by pol α. The DNA polymerase from bacteriophage RB69 has served as a model for eukaryotic B family polymerases for some time. The recent crystal structures of pol δ, α, and ε revealed similarities but also a number of unexpected differences between the eukaryotic polymerases and their bacteriophage counterpart, and also among the three yeast polymerases. This review will focus on their shared structural elements as well as the features that are unique to each of these polymerases.
Keywords: B family; DNA polymerase; eukaryotic replication; fidelity; proofreading.
Figures

Similar articles
-
In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta.Genetics. 2001 Sep;159(1):47-64. doi: 10.1093/genetics/159.1.47. Genetics. 2001. PMID: 11560886 Free PMC article.
-
Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors.Biol Direct. 2009 Mar 18;4:11. doi: 10.1186/1745-6150-4-11. Biol Direct. 2009. PMID: 19296856 Free PMC article.
-
Genetic Networks Required to Coordinate Chromosome Replication by DNA Polymerases α, δ, and ε in Saccharomyces cerevisiae.G3 (Bethesda). 2015 Aug 21;5(10):2187-97. doi: 10.1534/g3.115.021493. G3 (Bethesda). 2015. PMID: 26297725 Free PMC article.
-
Opportunities for new studies of nuclear DNA replication enzymology in budding yeast.Curr Genet. 2020 Apr;66(2):299-302. doi: 10.1007/s00294-019-01023-4. Epub 2019 Sep 6. Curr Genet. 2020. PMID: 31493018 Free PMC article. Review.
-
Reconstitution of a eukaryotic replisome reveals the mechanism of asymmetric distribution of DNA polymerases.Nucleus. 2016 Jul 3;7(4):360-8. doi: 10.1080/19491034.2016.1205774. Nucleus. 2016. PMID: 27416113 Free PMC article. Review.
Cited by
-
Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.Nat Commun. 2023 Dec 14;14(1):8306. doi: 10.1038/s41467-023-44125-x. Nat Commun. 2023. PMID: 38097591 Free PMC article.
-
Structures of human primosome elongation complexes.Nat Struct Mol Biol. 2023 May;30(5):579-583. doi: 10.1038/s41594-023-00971-3. Epub 2023 Apr 17. Nat Struct Mol Biol. 2023. PMID: 37069376 Free PMC article.
-
Identification of probable inhibitors for the DNA polymerase of the Monkeypox virus through the virtual screening approach.Int J Biol Macromol. 2023 Feb 28;229:515-528. doi: 10.1016/j.ijbiomac.2022.12.252. Epub 2022 Dec 28. Int J Biol Macromol. 2023. PMID: 36584781 Free PMC article.
-
Distinct clinical pattern of colorectal cancer patients with POLE mutations: A retrospective study on real-world data.Front Genet. 2022 Nov 21;13:963964. doi: 10.3389/fgene.2022.963964. eCollection 2022. Front Genet. 2022. PMID: 36479248 Free PMC article.
-
Genome Protection by DNA Polymerase θ.Annu Rev Genet. 2022 Nov 30;56:207-228. doi: 10.1146/annurev-genet-072920-041046. Epub 2022 Aug 26. Annu Rev Genet. 2022. PMID: 36028228 Free PMC article. Review.
References
-
- Beechem J. M., Otto M. R., Bloom L. B., Eritja R., Reha-Krantz L. J., Goodman M. F. (1998). Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry 37, 10144–10155 10.1021/bi980074b - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials


