Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers
- PMID: 24913235
- PMCID: PMC4076581
- DOI: 10.1084/jem.20131666
Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers
Abstract
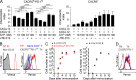
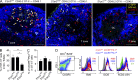
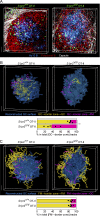
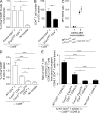
Follicular helper T (Tfh) cells access the B cell follicle to promote antibody responses and are particularly important for germinal center (GC) reactions. However, the molecular mechanisms of how Tfh cells are physically associated with GCs are incompletely understood. We report that the sphingosine-1-phosphate receptor 2 (S1PR2) gene is highly expressed in a subpopulation of Tfh cells that localizes in GCs. S1PR2-deficient Tfh cells exhibited reduced accumulation in GCs due to their impaired retention. T cells deficient in both S1PR2 and CXCR5 were ineffective in supporting GC responses compared with T cells deficient only in CXCR5. These results suggest that S1PR2 and CXCR5 cooperatively regulate localization of Tfh cells in GCs to support GC responses.
© 2014 Moriyama et al.
Figures

Similar articles
-
Primary germinal center-resident T follicular helper cells are a physiologically distinct subset of CXCR5hiPD-1hi T follicular helper cells.Immunity. 2022 Feb 8;55(2):272-289.e7. doi: 10.1016/j.immuni.2021.12.015. Epub 2022 Jan 25. Immunity. 2022. PMID: 35081372 Free PMC article.
-
Human T follicular helper cells: development and subsets.Adv Exp Med Biol. 2013;785:87-94. doi: 10.1007/978-1-4614-6217-0_10. Adv Exp Med Biol. 2013. PMID: 23456841 Review.
-
A distinct subpopulation of CD25- T-follicular regulatory cells localizes in the germinal centers.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):E6400-E6409. doi: 10.1073/pnas.1705551114. Epub 2017 Jul 11. Proc Natl Acad Sci U S A. 2017. PMID: 28698369 Free PMC article.
-
Induction of activated T follicular helper cells is critical for anti-FVIII inhibitor development in hemophilia A mice.Blood Adv. 2019 Oct 22;3(20):3099-3110. doi: 10.1182/bloodadvances.2019000650. Blood Adv. 2019. PMID: 31648333 Free PMC article.
-
Transcriptional regulation of follicular T-helper (Tfh) cells.Immunol Rev. 2013 Mar;252(1):139-45. doi: 10.1111/imr.12040. Immunol Rev. 2013. PMID: 23405901 Free PMC article. Review.
Cited by
-
Interleukin 9 mediates T follicular helper cell activation to promote antibody responses.Front Immunol. 2024 Sep 30;15:1441407. doi: 10.3389/fimmu.2024.1441407. eCollection 2024. Front Immunol. 2024. PMID: 39403384 Free PMC article.
-
Sphingosine-1-Phosphate Receptor 4 links neutrophils and early local inflammation to lymphocyte recruitment into the draining lymph node to facilitate robust germinal center formation.Front Immunol. 2024 Aug 12;15:1427509. doi: 10.3389/fimmu.2024.1427509. eCollection 2024. Front Immunol. 2024. PMID: 39188715 Free PMC article.
-
Specialized Tfh cell subsets driving type-1 and type-2 humoral responses in lymphoid tissue.Cell Discov. 2024 Jun 4;10(1):64. doi: 10.1038/s41421-024-00681-0. Cell Discov. 2024. PMID: 38834551 Free PMC article.
-
Transcriptional regulation of Tfh dynamics and the formation of immunological synapses.Exp Mol Med. 2024 Jun;56(6):1365-1372. doi: 10.1038/s12276-024-01254-7. Epub 2024 Jun 3. Exp Mol Med. 2024. PMID: 38825646 Free PMC article. Review.
-
Identification of Gα12-vs-Gα13-coupling determinants and development of a Gα12/13-coupled designer GPCR.Sci Rep. 2024 May 15;14(1):11119. doi: 10.1038/s41598-024-61506-4. Sci Rep. 2024. PMID: 38750247 Free PMC article.
References
-
- Cattoretti G., Mandelbaum J., Lee N., Chaves A.H., Mahler A.M., Chadburn A., Dalla-Favera R., Pasqualucci L., MacLennan A.J. 2009. Targeted disruption of the S1P2 sphingosine 1-phosphate receptor gene leads to diffuse large B-cell lymphoma formation. Cancer Res. 69:8686–8692 10.1158/0008-5472.CAN-09-1110 - DOI - PMC - PubMed
-
- Du W., Takuwa N., Yoshioka K., Okamoto Y., Gonda K., Sugihara K., Fukamizu A., Asano M., Takuwa Y. 2010. S1P2, the G protein-coupled receptor for sphingosine-1-phosphate, negatively regulates tumor angiogenesis and tumor growth in vivo in mice. Cancer Res. 70:772–781 10.1158/0008-5472.CAN-09-2722 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous


