Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination
- PMID: 24671993
- PMCID: PMC3965777
- DOI: 10.1523/JNEUROSCI.4314-13.2014
Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination
Abstract
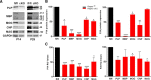
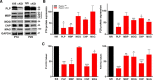
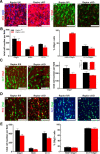
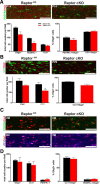
During CNS development, oligodendrocytes, the myelinating glia of the CNS, progress through multiple transitory stages before terminating into fully mature cells. Oligodendrocyte differentiation and myelination is a tightly regulated process requiring extracellular signals to converge to elicit specific translational and transcriptional changes. Our lab has previously shown that the protein kinases, Akt and mammalian Target of Rapamycin (mTOR), are important regulators of CNS myelination in vivo. mTOR functions through two distinct complexes, mTOR complex 1 (mTORC1) and mTORC2, by binding to either Raptor or Rictor, respectively. To establish whether the impact of mTOR on CNS myelination results from unique functions of mTORC1 or mTORC2 during CNS myelination, we conditionally ablated either Raptor or Rictor in the oligodendrocyte lineage, in vivo. We show that Raptor (mTORC1) is a positive regulator of developmental CNS mouse myelination when mTORC2 is functional, whereas Rictor (mTORC2) ablation has a modest positive effect on oligodendrocyte differentiation, and very little effect on myelination, when mTORC1 is functional. Also, we show that loss of Raptor in oligodendrocytes results in differential dysmyelination in specific areas of the CNS, with the greatest impact on spinal cord myelination.
Keywords: Raptor; Rictor; mTOR; oligodendrocyte.
Figures

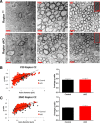
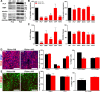
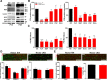
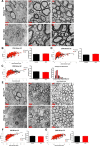
Similar articles
-
mTORC2 Loss in Oligodendrocyte Progenitor Cells Results in Regional Hypomyelination in the Central Nervous System.J Neurosci. 2023 Jan 25;43(4):540-558. doi: 10.1523/JNEUROSCI.0010-22.2022. Epub 2022 Dec 2. J Neurosci. 2023. PMID: 36460463 Free PMC article.
-
Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination.J Neurosci. 2014 Mar 26;34(13):4453-65. doi: 10.1523/JNEUROSCI.4311-13.2014. J Neurosci. 2014. PMID: 24671992 Free PMC article.
-
Balanced mTORC1 activity in oligodendrocytes is required for accurate CNS myelination.J Neurosci. 2014 Jun 18;34(25):8432-48. doi: 10.1523/JNEUROSCI.1105-14.2014. J Neurosci. 2014. PMID: 24948799 Free PMC article.
-
Regulators of Oligodendrocyte Differentiation.Cold Spring Harb Perspect Biol. 2024 Jun 3;16(6):a041358. doi: 10.1101/cshperspect.a041358. Cold Spring Harb Perspect Biol. 2024. PMID: 38503504 Review.
-
Purinergic signaling in oligodendrocyte development and function.J Neurochem. 2018 Apr;145(1):6-18. doi: 10.1111/jnc.14315. Epub 2018 Mar 25. J Neurochem. 2018. PMID: 29377124 Free PMC article. Review.
Cited by
-
The role of TSC1 and TSC2 proteins in neuronal axons.Mol Psychiatry. 2024 Apr;29(4):1165-1178. doi: 10.1038/s41380-023-02402-7. Epub 2024 Jan 11. Mol Psychiatry. 2024. PMID: 38212374 Review.
-
The Roles of Caloric Restriction Mimetics in Central Nervous System Demyelination and Remyelination.Curr Issues Mol Biol. 2023 Nov 27;45(12):9526-9548. doi: 10.3390/cimb45120596. Curr Issues Mol Biol. 2023. PMID: 38132442 Free PMC article. Review.
-
Glutamate delta-1 receptor regulates oligodendrocyte progenitor cell differentiation and myelination in normal and demyelinating conditions.PLoS One. 2023 Nov 20;18(11):e0294583. doi: 10.1371/journal.pone.0294583. eCollection 2023. PLoS One. 2023. PMID: 37983226 Free PMC article.
-
The AMPK activator metformin improves recovery from demyelination by shifting oligodendrocyte bioenergetics and accelerating OPC differentiation.Front Cell Neurosci. 2023 Oct 12;17:1254303. doi: 10.3389/fncel.2023.1254303. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37904733 Free PMC article.
-
Multifaceted role of mTOR (mammalian target of rapamycin) signaling pathway in human health and disease.Signal Transduct Target Ther. 2023 Oct 2;8(1):375. doi: 10.1038/s41392-023-01608-z. Signal Transduct Target Ther. 2023. PMID: 37779156 Free PMC article. Review.
References
-
- Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Rüegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

