Restriction of the conformational dynamics of the cyclic acyldepsipeptide antibiotics improves their antibacterial activity
- PMID: 24422534
- PMCID: PMC4004210
- DOI: 10.1021/ja410385c
Restriction of the conformational dynamics of the cyclic acyldepsipeptide antibiotics improves their antibacterial activity
Abstract
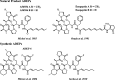
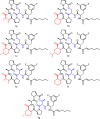
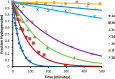
The cyclic acyldepsipeptide (ADEP) antibiotics are a new class of antibacterial agents that kill bacteria via a mechanism that is distinct from all clinically used drugs. These molecules bind and dysregulate the activity of the ClpP peptidase. The potential of these antibiotics as antibacterial drugs has been enhanced by the elimination of pharmacological liabilities through medicinal chemistry efforts. Here, we demonstrate that the ADEP conformation observed in the ADEP-ClpP crystal structure is fortified by transannular hydrogen bonding and can be further stabilized by judicious replacement of constituent amino acids within the peptidolactone core structure with more conformationally constrained counterparts. Evidence supporting constraint of the molecule into the bioactive conformer was obtained by measurements of deuterium-exchange kinetics of hydrogens that were proposed to be engaged in transannular hydrogen bonds. We show that the rigidified ADEP analogs bind and activate ClpP at lower concentrations in vitro. Remarkably, these compounds have up to 1200-fold enhanced antibacterial activity when compared to those with the peptidolactone core structure common to two ADEP natural products. This study compellingly demonstrates how rational modulation of conformational dynamics may be used to improve the bioactivities of natural products.
Figures
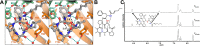
Similar articles
-
Diversity-oriented synthesis of cyclic acyldepsipeptides leads to the discovery of a potent antibacterial agent.Bioorg Med Chem. 2010 Oct 15;18(20):7193-202. doi: 10.1016/j.bmc.2010.08.032. Epub 2010 Aug 19. Bioorg Med Chem. 2010. PMID: 20833054
-
Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.Chembiochem. 2020 Jul 16;21(14):1997-2012. doi: 10.1002/cbic.201900787. Epub 2020 Apr 9. Chembiochem. 2020. PMID: 32181548 Free PMC article.
-
Total synthesis and antibacterial testing of the A54556 cyclic acyldepsipeptides isolated from Streptomyces hawaiiensis.J Nat Prod. 2014 Oct 24;77(10):2170-81. doi: 10.1021/np500158q. Epub 2014 Sep 25. J Nat Prod. 2014. PMID: 25255326
-
Acyldepsipeptide antibiotics--current state of knowledge.Pol J Microbiol. 2015;64(2):85-92. Pol J Microbiol. 2015. PMID: 26373166 Review.
-
The development of small-molecule modulators for ClpP protease activity.Mol Biosyst. 2016 Dec 20;13(1):23-31. doi: 10.1039/c6mb00644b. Mol Biosyst. 2016. PMID: 27831584 Review.
Cited by
-
Illuminating the multiple Lewis acidity of triaryl-boranes via atropisomeric dative adducts.Chem Sci. 2024 Aug 26;15(38):15679-89. doi: 10.1039/d4sc00925h. Online ahead of print. Chem Sci. 2024. PMID: 39257854 Free PMC article.
-
Protein degradation by a component of the chaperonin-linked protease ClpP.Genes Cells. 2024 Sep;29(9):695-709. doi: 10.1111/gtc.13141. Epub 2024 Jul 4. Genes Cells. 2024. PMID: 38965067 Free PMC article. Review.
-
One-pot chemoenzymatic syntheses of non-canonical amino acids.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae005. doi: 10.1093/jimb/kuae005. J Ind Microbiol Biotechnol. 2024. PMID: 38271597 Free PMC article. Review.
-
Identification of macrocyclic peptides which activate bacterial cylindrical proteases.RSC Med Chem. 2023 May 17;14(6):1186-1191. doi: 10.1039/d3md00136a. eCollection 2023 Jun 22. RSC Med Chem. 2023. PMID: 37360394 Free PMC article.
-
Combatting persister cells: The daunting task in post-antibiotics era.Cell Insight. 2023 Apr 24;2(4):100104. doi: 10.1016/j.cellin.2023.100104. eCollection 2023 Aug. Cell Insight. 2023. PMID: 37304393 Free PMC article. Review.
References
-
- Maurizi M. R.; Thompson M. W.; Singh S. K.; Kim S. Methods Enzymol. 1993, 244, 314–331. - PubMed
-
- Gominet M.; Seghezzi N.; Mazodier P.. Microbiology 2011, 157, 2226–2234. - PubMed
-
- Gottesman S.; Wickner S.; Maurizi M. R. Genes Dev. 1997, 11, 815–823. - PubMed
-
- Sauer R. T.; Baker T. A. Annu. Rev. Biochem. 2011, 80, 587–612. - PubMed
-
- Yu A. Y. H.; Houry W. A. FEBS Lett. 2007, 581, 3749–3757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical


