CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering
- PMID: 23907171
- PMCID: PMC3818127
- DOI: 10.1038/nbt.2675
CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering
Abstract
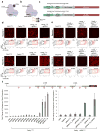
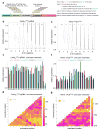
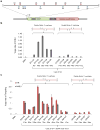
Prokaryotic type II CRISPR-Cas systems can be adapted to enable targeted genome modifications across a range of eukaryotes. Here we engineer this system to enable RNA-guided genome regulation in human cells by tethering transcriptional activation domains either directly to a nuclease-null Cas9 protein or to an aptamer-modified single guide RNA (sgRNA). Using this functionality we developed a transcriptional activation-based assay to determine the landscape of off-target binding of sgRNA:Cas9 complexes and compared it with the off-target activity of transcription activator-like (TALs) effectors. Our results reveal that specificity profiles are sgRNA dependent, and that sgRNA:Cas9 complexes and 18-mer TAL effectors can potentially tolerate 1-3 and 1-2 target mismatches, respectively. By engineering a requirement for cooperativity through offset nicking for genome editing or through multiple synergistic sgRNAs for robust transcriptional activation, we suggest methods to mitigate off-target phenomena. Our results expand the versatility of the sgRNA:Cas9 tool and highlight the critical need to engineer improved specificity.
Figures
Comment in
-
Staying on target with CRISPR-Cas.Nat Biotechnol. 2013 Sep;31(9):807-9. doi: 10.1038/nbt.2684. Nat Biotechnol. 2013. PMID: 24022156 No abstract available.
Similar articles
-
Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases.Genome Res. 2014 Jan;24(1):132-41. doi: 10.1101/gr.162339.113. Epub 2013 Nov 19. Genome Res. 2014. PMID: 24253446 Free PMC article.
-
CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences.Nucleic Acids Res. 2014 Jun;42(11):7473-85. doi: 10.1093/nar/gku402. Epub 2014 May 16. Nucleic Acids Res. 2014. PMID: 24838573 Free PMC article.
-
Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector.Nucleic Acids Res. 2014 Oct 29;42(19):e147. doi: 10.1093/nar/gku749. Epub 2014 Aug 13. Nucleic Acids Res. 2014. PMID: 25122746 Free PMC article.
-
CRISPR/Cas9 system as an innovative genetic engineering tool: Enhancements in sequence specificity and delivery methods.Biochim Biophys Acta. 2015 Dec;1856(2):234-43. doi: 10.1016/j.bbcan.2015.09.003. Epub 2015 Nov 11. Biochim Biophys Acta. 2015. PMID: 26434948 Review.
-
Application of CRISPR/Cas9 genome editing to the study and treatment of disease.Arch Toxicol. 2015 Jul;89(7):1023-34. doi: 10.1007/s00204-015-1504-y. Epub 2015 Apr 1. Arch Toxicol. 2015. PMID: 25827103 Review.
Cited by
-
dCas-Based Tools to Visualize Chromatin or Modify Epigenetic Marks at Specific Plant Genomic Loci.Methods Mol Biol. 2025;2873:305-332. doi: 10.1007/978-1-0716-4228-3_17. Methods Mol Biol. 2025. PMID: 39576609 Review.
-
Engineered PsCas9 enables therapeutic genome editing in mouse liver with lipid nanoparticles.Nat Commun. 2024 Nov 7;15(1):9173. doi: 10.1038/s41467-024-53418-8. Nat Commun. 2024. PMID: 39511150 Free PMC article.
-
CRISPR-Cas9-mediated homology-directed repair for precise gene editing.Mol Ther Nucleic Acids. 2024 Sep 26;35(4):102344. doi: 10.1016/j.omtn.2024.102344. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39494147 Free PMC article. Review.
-
Development of compact transcriptional effectors using high-throughput measurements in diverse contexts.Nat Biotechnol. 2024 Nov 1. doi: 10.1038/s41587-024-02442-6. Online ahead of print. Nat Biotechnol. 2024. PMID: 39487265
-
The potential of HBV cure: an overview of CRISPR-mediated HBV gene disruption.Front Genome Ed. 2024 Oct 9;6:1467449. doi: 10.3389/fgeed.2024.1467449. eCollection 2024. Front Genome Ed. 2024. PMID: 39444780 Free PMC article. Review.
References
-
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous


