Effects of histidine protonation and rotameric states on virtual screening of M. tuberculosis RmlC
- PMID: 23579613
- PMCID: PMC3639364
- DOI: 10.1007/s10822-013-9643-9
Effects of histidine protonation and rotameric states on virtual screening of M. tuberculosis RmlC
Abstract
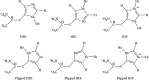
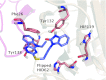
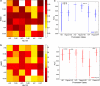
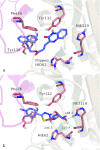
While it is well established that protonation and tautomeric states of ligands can significantly affect the results of virtual screening, such effects of ionizable residues of protein receptors are less well understood. In this study, we focus on histidine protonation and rotameric states and their impact on virtual screening of Mycobacterium tuberculosis enzyme RmlC. Depending on the net charge and the location of proton(s), a histidine can adopt three states: HIP (+1 charged, both δ- and ε-nitrogens protonated), HID (neutral, δ-nitrogen protonated), and HIE (neutral, ε-nitrogen protonated). Due to common ambiguities in X-ray crystal structures, a histidine may also be resolved as three additional states with its imidazole ring flipped. Here, we systematically investigate the predictive power of 36 receptor models with different protonation and rotameric states of two histidines in the RmlC active site by using results from a previous high-throughput screening. By measuring enrichment factors and area under the receiver operating characteristic curves, we show that virtual screening results vary depending on hydrogen bonding networks provided by the histidines, even in the cases where the ligand does not obviously interact with the side chain. Our results also suggest that, even with the help of widely used pKa prediction software, assigning histidine protonation and rotameric states for virtual screening can still be challenging and requires further examination and systematic characterization of the receptor-ligand complex.
Figures
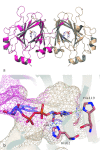
Similar articles
-
Virtual screening for the identification of novel inhibitors of Mycobacterium tuberculosis cell wall synthesis: inhibitors targeting RmlB and RmlC.Comput Biol Med. 2015 Mar;58:110-7. doi: 10.1016/j.compbiomed.2014.12.020. Epub 2015 Jan 5. Comput Biol Med. 2015. PMID: 25637777
-
Mycobacterium tuberculosis RmlC epimerase (Rv3465): a promising drug-target structure in the rhamnose pathway.Acta Crystallogr D Biol Crystallogr. 2004 May;60(Pt 5):895-902. doi: 10.1107/S0907444904005323. Epub 2004 Apr 21. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 15103135
-
Peptide binding to HLA-DP proteins at pH 5.0 and pH 7.0: a quantitative molecular docking study.BMC Struct Biol. 2012 Aug 5;12:20. doi: 10.1186/1472-6807-12-20. BMC Struct Biol. 2012. PMID: 22862845 Free PMC article.
-
The role of protonation states in ligand-receptor recognition and binding.Curr Pharm Des. 2013;19(23):4182-90. doi: 10.2174/1381612811319230004. Curr Pharm Des. 2013. PMID: 23170880 Free PMC article. Review.
-
Protonation and pK changes in protein-ligand binding.Q Rev Biophys. 2013 May;46(2):181-209. doi: 10.1017/S0033583513000024. Q Rev Biophys. 2013. PMID: 23889892 Free PMC article. Review.
Cited by
-
Identification of echinoderm metabolites as potential inhibitors targeting wild-type and mutant forms of Escherichia coli RNA polymerase (RpoB) for tuberculosis treatment.PLoS One. 2024 Aug 30;19(8):e0304587. doi: 10.1371/journal.pone.0304587. eCollection 2024. PLoS One. 2024. PMID: 39213289 Free PMC article.
-
Insilico exploration C. koseri ATP synthase inhibitors by pharmacophore-based virtual screening, molecular docking and MD simulation.PLoS One. 2024 Aug 22;19(8):e0308251. doi: 10.1371/journal.pone.0308251. eCollection 2024. PLoS One. 2024. PMID: 39173004 Free PMC article.
-
Structure-based virtual identification of natural inhibitors of SARS-CoV-2 and its Delta and Omicron variant proteins.Future Virol. 2023 May;18(7):421-438. doi: 10.2217/fvl-2022-0184. Epub 2023 Jun 1. Future Virol. 2023. PMID: 38051986 Free PMC article.
-
In silico identification of deep-sea fungal alkaloids as potential inhibitors of SARS-CoV-2, Delta and Omicron spikes.Future Virol. 2023 Oct:10.2217/fvl-2023-0102. doi: 10.2217/fvl-2023-0102. Epub 2023 Oct 26. Future Virol. 2023. PMID: 37908844 Free PMC article.
-
Curcumin nanoparticles: physicochemical fabrication, characterization, antioxidant, enzyme inhibition, molecular docking and simulation studies.RSC Adv. 2023 Jul 24;13(32):22268-22280. doi: 10.1039/d3ra01432k. eCollection 2023 Jul 19. RSC Adv. 2023. PMID: 37492507 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


