An integrated map of genetic variation from 1,092 human genomes
- PMID: 23128226
- PMCID: PMC3498066
- DOI: 10.1038/nature11632
An integrated map of genetic variation from 1,092 human genomes
Abstract
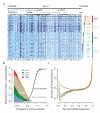
By characterizing the geographic and functional spectrum of human genetic variation, the 1000 Genomes Project aims to build a resource to help to understand the genetic contribution to disease. Here we describe the genomes of 1,092 individuals from 14 populations, constructed using a combination of low-coverage whole-genome and exome sequencing. By developing methods to integrate information across several algorithms and diverse data sources, we provide a validated haplotype map of 38 million single nucleotide polymorphisms, 1.4 million short insertions and deletions, and more than 14,000 larger deletions. We show that individuals from different populations carry different profiles of rare and common variants, and that low-frequency variants show substantial geographic differentiation, which is further increased by the action of purifying selection. We show that evolutionary conservation and coding consequence are key determinants of the strength of purifying selection, that rare-variant load varies substantially across biological pathways, and that each individual contains hundreds of rare non-coding variants at conserved sites, such as motif-disrupting changes in transcription-factor-binding sites. This resource, which captures up to 98% of accessible single nucleotide polymorphisms at a frequency of 1% in related populations, enables analysis of common and low-frequency variants in individuals from diverse, including admixed, populations.
Figures


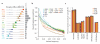


Comment in
-
A new era of human population genetics.Genome Biol. 2012 Dec 26;13(12):182. doi: 10.1186/gb-2012-13-12-182. Genome Biol. 2012. PMID: 23268745 Free PMC article.
Similar articles
-
A map of human genome variation from population-scale sequencing.Nature. 2010 Oct 28;467(7319):1061-73. doi: 10.1038/nature09534. Nature. 2010. PMID: 20981092 Free PMC article.
-
A global reference for human genetic variation.Nature. 2015 Oct 1;526(7571):68-74. doi: 10.1038/nature15393. Nature. 2015. PMID: 26432245 Free PMC article.
-
An integrated map of structural variation in 2,504 human genomes.Nature. 2015 Oct 1;526(7571):75-81. doi: 10.1038/nature15394. Nature. 2015. PMID: 26432246 Free PMC article.
-
Small insertions and deletions (INDELs) in human genomes.Hum Mol Genet. 2010 Oct 15;19(R2):R131-6. doi: 10.1093/hmg/ddq400. Epub 2010 Sep 21. Hum Mol Genet. 2010. PMID: 20858594 Free PMC article. Review.
-
Molecular genetic studies of complex phenotypes.Transl Res. 2012 Feb;159(2):64-79. doi: 10.1016/j.trsl.2011.08.001. Epub 2011 Aug 31. Transl Res. 2012. PMID: 22243791 Free PMC article. Review.
Cited by
-
Genomic and transcriptomic analyses identify distinctive features of triple-negative inflammatory breast cancer.NPJ Precis Oncol. 2024 Nov 18;8(1):265. doi: 10.1038/s41698-024-00729-0. NPJ Precis Oncol. 2024. PMID: 39558017 Free PMC article.
-
GenoSiS: A Biobank-Scale Genotype Similarity Search Architecture for Creating Dynamic Patient-Match Cohorts.bioRxiv [Preprint]. 2024 Nov 3:2024.11.02.621671. doi: 10.1101/2024.11.02.621671. bioRxiv. 2024. PMID: 39554195 Free PMC article. Preprint.
-
DNA methylation biomarkers and myopia: a multi-omics study integrating GWAS, mQTL and eQTL data.Clin Epigenetics. 2024 Nov 13;16(1):157. doi: 10.1186/s13148-024-01772-1. Clin Epigenetics. 2024. PMID: 39538342 Free PMC article.
-
Fine-mapping and molecular characterisation of primary sclerosing cholangitis genetic risk loci.Nat Commun. 2024 Nov 6;15(1):9594. doi: 10.1038/s41467-024-53602-w. Nat Commun. 2024. PMID: 39505854 Free PMC article.
-
The potential role of next-generation sequencing in identifying MET amplification and disclosing resistance mechanisms in NSCLC patients with osimertinib resistance.Front Oncol. 2024 Oct 21;14:1470827. doi: 10.3389/fonc.2024.1470827. eCollection 2024. Front Oncol. 2024. PMID: 39497720 Free PMC article.
References
-
- Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi:10.1126/science.1181498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01HG5208/HG/NHGRI NIH HHS/United States
- T32 HL094284/HL/NHLBI NIH HHS/United States
- T32GM7748/GM/NIGMS NIH HHS/United States
- UL1RR024131/RR/NCRR NIH HHS/United States
- R01HG4960/HG/NHGRI NIH HHS/United States
- R01 HG002898/HG/NHGRI NIH HHS/United States
- R01 ES015794/ES/NIEHS NIH HHS/United States
- U01HG5725/HG/NHGRI NIH HHS/United States
- RC2HG5581/HG/NHGRI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- T15 LM007056/LM/NLM NIH HHS/United States
- AI077439/AI/NIAID NIH HHS/United States
- R01 MH084698/MH/NIMH NIH HHS/United States
- RG/09/012/28096/BHF_/British Heart Foundation/United Kingdom
- R01 HG004960/HG/NHGRI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U01 HG005208/HG/NHGRI NIH HHS/United States
- AI2009061/AI/NIAID NIH HHS/United States
- ES015794/ES/NIEHS NIH HHS/United States
- R01 HG004719/HG/NHGRI NIH HHS/United States
- G0900747(91070)/MRC_/Medical Research Council/United Kingdom
- P41HG2371/HG/NHGRI NIH HHS/United States
- R01HG2898/HG/NHGRI NIH HHS/United States
- DP2OD6514/OD/NIH HHS/United States
- WT085475/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- R01HG3698/HG/NHGRI NIH HHS/United States
- RC2 HG005552/HG/NHGRI NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- RC2HL102925/HL/NHLBI NIH HHS/United States
- U01HG5715/HG/NHGRI NIH HHS/United States
- R01GM59290/GM/NIGMS NIH HHS/United States
- R01HL95045/HL/NHLBI NIH HHS/United States
- U41HG4568/HG/NHGRI NIH HHS/United States
- U01 HG005211/HG/NHGRI NIH HHS/United States
- U01 HG005214/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HG006569/HG/NHGRI NIH HHS/United States
- P41 HG002371/HG/NHGRI NIH HHS/United States
- R01 HL078885/HL/NHLBI NIH HHS/United States
- UL1 RR024131/RR/NCRR NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- T32 GM008283/GM/NIGMS NIH HHS/United States
- U01 HG005728/HG/NHGRI NIH HHS/United States
- U01HG6569/HG/NHGRI NIH HHS/United States
- WT085532AIA/WT_/Wellcome Trust/United Kingdom
- 096599/WT_/Wellcome Trust/United Kingdom
- U01 HG005715/HG/NHGRI NIH HHS/United States
- WT090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- G0900747/MRC_/Medical Research Council/United Kingdom
- BB/I02593X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U01HG5211/HG/NHGRI NIH HHS/United States
- U01HG6513/HG/NHGRI NIH HHS/United States
- G12 MD007579/MD/NIMHD NIH HHS/United States
- U54HG3079/HG/NHGRI NIH HHS/United States
- G0801823/MRC_/Medical Research Council/United Kingdom
- HL078885/HL/NHLBI NIH HHS/United States
- P20 MD006899/MD/NIMHD NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- U01HG5214/HG/NHGRI NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- U41 HG004568/HG/NHGRI NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- R01 GM059290/GM/NIGMS NIH HHS/United States
- 095908/WT_/Wellcome Trust/United Kingdom
- R01MH84698/MH/NIMH NIH HHS/United States
- R01 HG005701/HG/NHGRI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U01 HG005725/HG/NHGRI NIH HHS/United States
- R01 CA166661/CA/NCI NIH HHS/United States
- U01HG5728/HG/NHGRI NIH HHS/United States
- HHSN268201100040C/HL/NHLBI NIH HHS/United States
- R01CA166661/CA/NCI NIH HHS/United States
- R01 HG003698/HG/NHGRI NIH HHS/United States
- T15LM7033/LM/NLM NIH HHS/United States
- RC2HG5552/HG/NHGRI NIH HHS/United States
- P01HG4120/HG/NHGRI NIH HHS/United States
- R01 HL095045/HL/NHLBI NIH HHS/United States
- U01HG5209/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- T32GM8283/GM/NIGMS NIH HHS/United States
- R01 HG007022/HG/NHGRI NIH HHS/United States
- U54HG3067/HG/NHGRI NIH HHS/United States
- 086084/WT_/Wellcome Trust/United Kingdom
- P41HG4221/HG/NHGRI NIH HHS/United States
- DP2 OD006514/OD/NIH HHS/United States
- RC2 HG005581/HG/NHGRI NIH HHS/United States
- 085532/WT_/Wellcome Trust/United Kingdom
- R01HG5701/HG/NHGRI NIH HHS/United States
- U01 HG005209/HG/NHGRI NIH HHS/United States
- T32HL94284/HL/NHLBI NIH HHS/United States
- R01 HL088133/HL/NHLBI NIH HHS/United States
- WT089250/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- P41 HG004221/HG/NHGRI NIH HHS/United States
- P01 HG004120/HG/NHGRI NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
- G0701805/MRC_/Medical Research Council/United Kingdom
- WT095552/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- R01HG4719/HG/NHGRI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- U54HG3273/HG/NHGRI NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
- RG/09/12/28096/BHF_/British Heart Foundation/United Kingdom
- WT086084/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- BB/I021213/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G12 RR003050/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources


