The date of interbreeding between Neandertals and modern humans
- PMID: 23055938
- PMCID: PMC3464203
- DOI: 10.1371/journal.pgen.1002947
The date of interbreeding between Neandertals and modern humans
Abstract
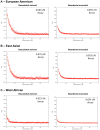
Comparisons of DNA sequences between Neandertals and present-day humans have shown that Neandertals share more genetic variants with non-Africans than with Africans. This could be due to interbreeding between Neandertals and modern humans when the two groups met subsequent to the emergence of modern humans outside Africa. However, it could also be due to population structure that antedates the origin of Neandertal ancestors in Africa. We measure the extent of linkage disequilibrium (LD) in the genomes of present-day Europeans and find that the last gene flow from Neandertals (or their relatives) into Europeans likely occurred 37,000-86,000 years before the present (BP), and most likely 47,000-65,000 years ago. This supports the recent interbreeding hypothesis and suggests that interbreeding may have occurred when modern humans carrying Upper Paleolithic technologies encountered Neandertals as they expanded out of Africa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
IBD Sharing between Africans, Neandertals, and Denisovans.Genome Biol Evol. 2016 Dec 1;8(12):3406-3416. doi: 10.1093/gbe/evw234. Genome Biol Evol. 2016. PMID: 28158547 Free PMC article.
-
No evidence of Neandertal admixture in the mitochondrial genomes of early European modern humans and contemporary Europeans.Am J Phys Anthropol. 2011 Oct;146(2):242-52. doi: 10.1002/ajpa.21569. Epub 2011 Aug 24. Am J Phys Anthropol. 2011. PMID: 21913172
-
A draft sequence of the Neandertal genome.Science. 2010 May 7;328(5979):710-722. doi: 10.1126/science.1188021. Science. 2010. PMID: 20448178 Free PMC article.
-
[Progresses on Neandertal genomics].Yi Chuan. 2012 Jun;34(6):659-65. doi: 10.3724/sp.j.1005.2012.00659. Yi Chuan. 2012. PMID: 22698735 Review. Chinese.
-
Neandertals revised.Proc Natl Acad Sci U S A. 2016 Jun 7;113(23):6372-9. doi: 10.1073/pnas.1521269113. Proc Natl Acad Sci U S A. 2016. PMID: 27274044 Free PMC article. Review.
Cited by
-
Life history and ancestry of the late Upper Palaeolithic infant from Grotta delle Mura, Italy.Nat Commun. 2024 Sep 20;15(1):8248. doi: 10.1038/s41467-024-51150-x. Nat Commun. 2024. PMID: 39304646 Free PMC article.
-
Global and Local Ancestry and its Importance: A Review.Curr Genomics. 2024;25(4):237-260. doi: 10.2174/0113892029298909240426094055. Epub 2024 May 9. Curr Genomics. 2024. PMID: 39156729 Free PMC article. Review.
-
Denisovan admixture facilitated environmental adaptation in Papua New Guinean populations.Proc Natl Acad Sci U S A. 2024 Jun 25;121(26):e2405889121. doi: 10.1073/pnas.2405889121. Epub 2024 Jun 18. Proc Natl Acad Sci U S A. 2024. PMID: 38889149 Free PMC article.
-
Neandertal ancestry through time: Insights from genomes of ancient and present-day humans.bioRxiv [Preprint]. 2024 May 13:2024.05.13.593955. doi: 10.1101/2024.05.13.593955. bioRxiv. 2024. PMID: 38798350 Free PMC article. Preprint.
-
Archaic Introgression Shaped Human Circadian Traits.Genome Biol Evol. 2023 Dec 1;15(12):evad203. doi: 10.1093/gbe/evad203. Genome Biol Evol. 2023. PMID: 38095367 Free PMC article.
References
-
- Krause J, Orlando L, Serre D, Viola B, Prufer K, et al. (2007) Neanderthals in central Asia and Siberia. Nature 449: 902–904. - PubMed
-
- Currat M, Excoffier L (2004) Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol 2: e421 doi:10.1371/journal.pbio.0020421 - DOI - PMC - PubMed
-
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, et al. (2009) Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325: 318–321. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous


