Four new bat species (Rhinolophus hildebrandtii complex) reflect Plio-Pleistocene divergence of dwarfs and giants across an Afromontane archipelago
- PMID: 22984399
- PMCID: PMC3440430
- DOI: 10.1371/journal.pone.0041744
Four new bat species (Rhinolophus hildebrandtii complex) reflect Plio-Pleistocene divergence of dwarfs and giants across an Afromontane archipelago
Abstract
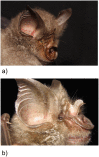
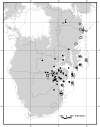
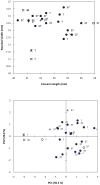
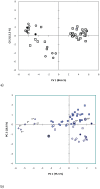
Gigantism and dwarfism evolve in vertebrates restricted to islands. We describe four new species in the Rhinolophus hildebrandtii species-complex of horseshoe bats, whose evolution has entailed adaptive shifts in body size. We postulate that vicissitudes of palaeoenvironments resulted in gigantism and dwarfism in habitat islands fragmented across eastern and southern Africa. Mitochondrial and nuclear DNA sequences recovered two clades of R. hildebrandtii senso lato which are paraphyletic with respect to a third lineage (R. eloquens). Lineages differ by 7.7 to 9.0% in cytochrome b sequences. Clade 1 includes R. hildebrandtii sensu stricto from the east African highlands and three additional vicariants that speciated across an Afromontane archipelago through the Plio-Pleistocene, extending from the Kenyan Highlands through the Eastern Arc, northern Mozambique and the Zambezi Escarpment to the eastern Great Escarpment of South Africa. Clade 2 comprises one species confined to lowland savanna habitats (Mozambique and Zimbabwe). A third clade comprises R. eloquens from East Africa. Speciation within Clade 1 is associated with fixed differences in echolocation call frequency, and cranial shape and size in populations isolated since the late Pliocene (ca 3.74 Mya). Relative to the intermediate-sized savanna population (Clade 2), these island-populations within Clade 1 are characterised by either gigantism (South African eastern Great Escarpment and Mts Mabu and Inago in Mozambique) or dwarfism (Lutope-Ngolangola Gorge, Zimbabwe and Soutpansberg Mountains, South Africa). Sympatry between divergent clades (Clade 1 and Clade 2) at Lutope-Ngolangola Gorge (NW Zimbabwe) is attributed to recent range expansions. We propose an "Allometric Speciation Hypothesis", which attributes the evolution of this species complex of bats to divergence in constant frequency (CF) sonar calls. The origin of species-specific peak frequencies (overall range = 32 to 46 kHz) represents the allometric effect of adaptive divergence in skull size, represented in the evolution of gigantism and dwarfism in habitat islands.
Conflict of interest statement
Figures





Similar articles
-
Southern Africa's Great Escarpment as an amphitheater of climate-driven diversification and a buffer against future climate change in bats.Glob Chang Biol. 2024 Jun;30(6):e17344. doi: 10.1111/gcb.17344. Glob Chang Biol. 2024. PMID: 38837566
-
Rapid and repeated origin of insular gigantism and dwarfism in Australian tiger snakes.Evolution. 2005 Jan;59(1):226-33. Evolution. 2005. PMID: 15792242
-
Molecular phylogenetics of the African horseshoe bats (Chiroptera: Rhinolophidae): expanded geographic and taxonomic sampling of the Afrotropics.BMC Evol Biol. 2019 Aug 22;19(1):166. doi: 10.1186/s12862-019-1485-1. BMC Evol Biol. 2019. PMID: 31434566 Free PMC article.
-
Phenotypic convergence in genetically distinct lineages of a Rhinolophus species complex (Mammalia, Chiroptera).PLoS One. 2013 Dec 3;8(12):e82614. doi: 10.1371/journal.pone.0082614. eCollection 2013. PLoS One. 2013. PMID: 24312666 Free PMC article.
-
The comparative phylogeography of fruit bats of the tribe Scotonycterini (Chiroptera, Pteropodidae) reveals cryptic species diversity related to African Pleistocene forest refugia.C R Biol. 2015 Mar;338(3):197-211. doi: 10.1016/j.crvi.2014.12.003. Epub 2015 Jan 27. C R Biol. 2015. PMID: 25636226
Cited by
-
An island in a sea of sand: a first checklist of the herpetofauna of the Serra da Neve inselberg, southwestern Angola.Zookeys. 2024 May 14;1201:167-217. doi: 10.3897/zookeys.1201.120750. eCollection 2024. Zookeys. 2024. PMID: 38779586 Free PMC article.
-
A biogeographical appraisal of the threatened South East Africa Montane Archipelago ecoregion.Sci Rep. 2024 Mar 12;14(1):5971. doi: 10.1038/s41598-024-54671-z. Sci Rep. 2024. PMID: 38472297 Free PMC article.
-
Unraveling the macroevolution of horseshoe bats (Chiroptera: Rhinolophidae: Rhinolophus).Zool Res. 2023 Jan 18;44(1):169-182. doi: 10.24272/j.issn.2095-8137.2022.265. Zool Res. 2023. PMID: 36579403 Free PMC article.
-
Does evolution of echolocation calls and morphology in Molossus result from convergence or stasis?PLoS One. 2020 Sep 24;15(9):e0238261. doi: 10.1371/journal.pone.0238261. eCollection 2020. PLoS One. 2020. PMID: 32970683 Free PMC article.
-
Comparative phylogeography of mainland and insular species of Neotropical molossid bats (Molossus).Ecol Evol. 2019 Dec 19;10(1):389-409. doi: 10.1002/ece3.5903. eCollection 2020 Jan. Ecol Evol. 2019. PMID: 31993120 Free PMC article.
References
-
- Van Valen LM (1973) Pattern and the balance of nature. Evolutionary Theory 1: 31–49.
-
- Lomolino MV (2005) Body size evolution in insular vertebrates: generality of the island rule. Journal of Biogeography 32: 1683–1699.
-
- Meiri S, Dayan T, Simberloff D (2006) The generality of the island rule re-examined. Journal of Biogeography 33: 1571–1577.
-
- Jacobs DS (1996) Morphological divergence in an insular bat, Lasiurus cinereus semotus . Functional Ecology 10: 622–630.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous


