Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana
- PMID: 22969436
- PMCID: PMC3435251
- DOI: 10.1371/journal.pgen.1002923
Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana
Abstract
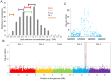
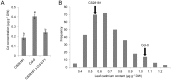
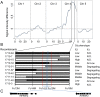
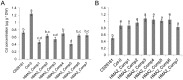
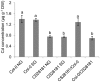
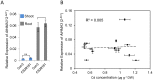
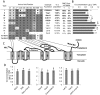
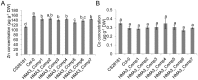
Understanding the mechanism of cadmium (Cd) accumulation in plants is important to help reduce its potential toxicity to both plants and humans through dietary and environmental exposure. Here, we report on a study to uncover the genetic basis underlying natural variation in Cd accumulation in a world-wide collection of 349 wild collected Arabidopsis thaliana accessions. We identified a 4-fold variation (0.5-2 µg Cd g(-1) dry weight) in leaf Cd accumulation when these accessions were grown in a controlled common garden. By combining genome-wide association mapping, linkage mapping in an experimental F2 population, and transgenic complementation, we reveal that HMA3 is the sole major locus responsible for the variation in leaf Cd accumulation we observe in this diverse population of A. thaliana accessions. Analysis of the predicted amino acid sequence of HMA3 from 149 A. thaliana accessions reveals the existence of 10 major natural protein haplotypes. Association of these haplotypes with leaf Cd accumulation and genetics complementation experiments indicate that 5 of these haplotypes are active and 5 are inactive, and that elevated leaf Cd accumulation is associated with the reduced function of HMA3 caused by a nonsense mutation and polymorphisms that change two specific amino acids.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots.Plant Physiol. 2012 Feb;158(2):790-800. doi: 10.1104/pp.111.190983. Epub 2011 Dec 19. Plant Physiol. 2012. PMID: 22184655 Free PMC article.
-
Variation in sulfur and selenium accumulation is controlled by naturally occurring isoforms of the key sulfur assimilation enzyme ADENOSINE 5'-PHOSPHOSULFATE REDUCTASE2 across the Arabidopsis species range.Plant Physiol. 2014 Nov;166(3):1593-608. doi: 10.1104/pp.114.247825. Epub 2014 Sep 22. Plant Physiol. 2014. PMID: 25245030 Free PMC article.
-
Genome-wide association analysis and QTL mapping reveal the genetic control of cadmium accumulation in maize leaf.BMC Genomics. 2018 Jan 25;19(1):91. doi: 10.1186/s12864-017-4395-x. BMC Genomics. 2018. PMID: 29370753 Free PMC article.
-
Association mapping of cadmium, copper and hydrogen peroxide tolerance of roots and translocation capacities of cadmium and copper in Arabidopsis thaliana.Physiol Plant. 2009 Nov;137(3):235-48. doi: 10.1111/j.1399-3054.2009.01286.x. Physiol Plant. 2009. PMID: 19832939
-
Exploiting natural variation to uncover candidate genes that control element accumulation in Arabidopsis thaliana.New Phytol. 2012 Mar;193(4):859-66. doi: 10.1111/j.1469-8137.2011.03977.x. New Phytol. 2012. PMID: 22403822 Review.
Cited by
-
A CC-Type Glutaredoxins GRX480 Functions in Cadmium Tolerance by Maintaining Redox Homeostasis in Arabidopsis.Int J Mol Sci. 2024 Oct 25;25(21):11455. doi: 10.3390/ijms252111455. Int J Mol Sci. 2024. PMID: 39519008 Free PMC article.
-
Genome-wide association analysis reveals genes controlling an antagonistic effect of biotic and osmotic stress on Arabidopsis thaliana growth.Mol Plant Pathol. 2024 Mar;25(3):e13436. doi: 10.1111/mpp.13436. Mol Plant Pathol. 2024. PMID: 38460112 Free PMC article.
-
The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.).Plant Cell Rep. 2024 Jan 22;43(2):44. doi: 10.1007/s00299-023-03112-8. Plant Cell Rep. 2024. PMID: 38246890
-
Genetic dissection of maize (Zea maysL.) trace element traits using genome-wide association studies.BMC Plant Biol. 2023 Dec 8;23(1):631. doi: 10.1186/s12870-023-04643-8. BMC Plant Biol. 2023. PMID: 38062375 Free PMC article.
-
Genome-wide association studies identify loci controlling specialized seed metabolites in Arabidopsis.Plant Physiol. 2024 Feb 29;194(3):1705-1721. doi: 10.1093/plphys/kiad511. Plant Physiol. 2024. PMID: 37758174 Free PMC article.
References
-
- Ursinyova M HV (2000) Cadmium in the environment of Central Europe. In: Markert Bernd A FK, editor. Trace Elements: their distribution and effects in the environment. 1 ed. Kindligton: Elsevier Science Ltd. pp. 87–108.
-
- Nawrot T, Plusquin M, Hogervorst J, Roels HA, Celis H, et al. (2006) Environmental exposure to cadmium and risk of cancer: a prospective population-based study. Lancet Oncol 7: 119–126. - PubMed
-
- Verbruggen N, Hermans C, Schat H (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol 12: 364–372. - PubMed
-
- Peralta-Videa JR, Lopez ML, Narayan M, Saupe G, Gardea-Torresdey J (2009) The biochemistry of environmental heavy metal uptake by plants: implications for the food chain. Int J Biochem Cell Biol 41: 1665–1677. - PubMed
-
- LeDuc DL, Terry N (2005) Phytoremediation of toxic trace elements in soil and water. J Ind Microbiol Biotechnol 32: 514–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous


