SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing
- PMID: 22506599
- PMCID: PMC3342519
- DOI: 10.1089/cmb.2012.0021
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing
Abstract
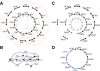
The lion's share of bacteria in various environments cannot be cloned in the laboratory and thus cannot be sequenced using existing technologies. A major goal of single-cell genomics is to complement gene-centric metagenomic data with whole-genome assemblies of uncultivated organisms. Assembly of single-cell data is challenging because of highly non-uniform read coverage as well as elevated levels of sequencing errors and chimeric reads. We describe SPAdes, a new assembler for both single-cell and standard (multicell) assembly, and demonstrate that it improves on the recently released E+V-SC assembler (specialized for single-cell data) and on popular assemblers Velvet and SoapDeNovo (for multicell data). SPAdes generates single-cell assemblies, providing information about genomes of uncultivatable bacteria that vastly exceeds what may be obtained via traditional metagenomics studies. SPAdes is available online ( http://bioinf.spbau.ru/spades ). It is distributed as open source software.
Figures




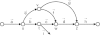
Similar articles
-
Assembling single-cell genomes and mini-metagenomes from chimeric MDA products.J Comput Biol. 2013 Oct;20(10):714-37. doi: 10.1089/cmb.2013.0084. J Comput Biol. 2013. PMID: 24093227 Free PMC article.
-
IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth.Bioinformatics. 2012 Jun 1;28(11):1420-8. doi: 10.1093/bioinformatics/bts174. Epub 2012 Apr 11. Bioinformatics. 2012. PMID: 22495754
-
Assembling short reads from jumping libraries with large insert sizes.Bioinformatics. 2015 Oct 15;31(20):3262-8. doi: 10.1093/bioinformatics/btv337. Epub 2015 Jun 3. Bioinformatics. 2015. PMID: 26040456
-
Single cell genome sequencing.Curr Opin Biotechnol. 2012 Jun;23(3):437-43. doi: 10.1016/j.copbio.2011.11.018. Epub 2011 Dec 7. Curr Opin Biotechnol. 2012. PMID: 22154471 Free PMC article. Review.
-
Metagenomic Data Assembly - The Way of Decoding Unknown Microorganisms.Front Microbiol. 2021 Mar 23;12:613791. doi: 10.3389/fmicb.2021.613791. eCollection 2021. Front Microbiol. 2021. PMID: 33833738 Free PMC article. Review.
Cited by
-
Genome assembly of an endemic butterfly (Minois Aurata) shed light on the genetic mechanisms underlying ecological adaptation to arid valley habitat.BMC Genomics. 2024 Nov 23;25(1):1134. doi: 10.1186/s12864-024-11058-8. BMC Genomics. 2024. PMID: 39580397 Free PMC article.
-
Phage cocktail amikacin combination as a potential therapy for bacteremia associated with carbapenemase producing colistin resistant Klebsiella pneumoniae.Sci Rep. 2024 Nov 22;14(1):28992. doi: 10.1038/s41598-024-79924-9. Sci Rep. 2024. PMID: 39578508 Free PMC article.
-
Methylocystis borbori sp.nov., a novel methanotrophic bacterium from the sludge of a freshwater lake and its metabolic properties.Antonie Van Leeuwenhoek. 2024 Nov 22;118(1):29. doi: 10.1007/s10482-024-02039-8. Antonie Van Leeuwenhoek. 2024. PMID: 39576297
-
The first complete chloroplast genome of Halodule uninervis (Forssk.) Boiss. 1882 (Cymodoceaceae).Mitochondrial DNA B Resour. 2024 Nov 20;9(11):1564-1568. doi: 10.1080/23802359.2024.2429635. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 39575205 Free PMC article.
-
The Complete Genome Sequences of 3 Species of Malacoctenus Fishes (Labrisomidae, Blenniiformes).Biodivers Genomes. 2024;2024:10.56179/001c.125783. doi: 10.56179/001c.125783. Epub 2024 Nov 10. Biodivers Genomes. 2024. PMID: 39574512 Free PMC article.
References
-
- Bandeira N. Clauser K. Pevzner P. Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol. Cell Proteomics. 2007;6:1123–1134. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


