Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium
- PMID: 21472017
- PMCID: PMC3176510
- DOI: 10.1038/ismej.2011.36
Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium
Abstract
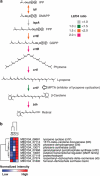
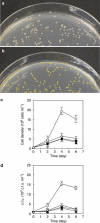
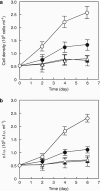
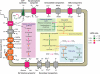
Proteorhodopsin (PR) is a photoprotein that functions as a light-driven proton pump in diverse marine Bacteria and Archaea. Recent studies have suggested that PR may enhance both growth rate and yield in some flavobacteria when grown under nutrient-limiting conditions in the light. The direct involvement of PR, and the metabolic details enabling light-stimulated growth, however, remain uncertain. Here, we surveyed transcriptional and growth responses of a PR-containing marine flavobacterium during carbon-limited growth in the light and the dark. As previously reported (Gómez-Consarnau et al., 2007), Dokdonia strain MED134 exhibited light-enhanced growth rates and cell yields under low carbon growth conditions. Inhibition of retinal biosynthesis abolished the light-stimulated growth response, supporting a direct role for retinal-bound PR in light-enhanced growth. Among protein-coding transcripts, both PR and retinal biosynthetic enzymes showed significant upregulation in the light. Other light-associated proteins, including bacterial cryptochrome and DNA photolyase, were also expressed at significantly higher levels in the light. Membrane transporters for Na(+)/phosphate and Na(+)/alanine symporters, and the Na(+)-translocating NADH-quinone oxidoreductase (NQR) linked electron transport chain, were also significantly upregulated in the light. Culture experiments using a specific inhibitor of Na(+)-translocating NQR indicated that sodium pumping via NQR is a critical metabolic process in the light-stimulated growth of MED134. In total, the results suggested the importance of both the PR-enabled, light-driven proton gradient, as well as the generation of a Na(+) ion gradient, as essential components for light-enhanced growth in these flavobacteria.
Figures
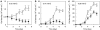
Similar articles
-
Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea.Appl Environ Microbiol. 2010 May;76(10):3187-97. doi: 10.1128/AEM.02971-09. Epub 2010 Mar 19. Appl Environ Microbiol. 2010. PMID: 20305030 Free PMC article.
-
Diversity and functional analysis of proteorhodopsin in marine Flavobacteria.Environ Microbiol. 2012 May;14(5):1240-8. doi: 10.1111/j.1462-2920.2012.02702.x. Epub 2012 Feb 13. Environ Microbiol. 2012. PMID: 22329552
-
Diversity and functional analysis of light-driven pumping rhodopsins in marine Flavobacteria.Microbiologyopen. 2016 Apr;5(2):212-23. doi: 10.1002/mbo3.321. Epub 2015 Dec 13. Microbiologyopen. 2016. PMID: 26663527 Free PMC article.
-
Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology.Microbiol Mol Biol Rev. 2016 Sep 14;80(4):929-54. doi: 10.1128/MMBR.00003-16. Print 2016 Dec. Microbiol Mol Biol Rev. 2016. PMID: 27630250 Free PMC article. Review.
-
Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump.Biochim Biophys Acta. 2010 Jun-Jul;1797(6-7):738-46. doi: 10.1016/j.bbabio.2009.12.020. Epub 2010 Jan 4. Biochim Biophys Acta. 2010. PMID: 20056102 Review.
Cited by
-
Niche differentiation within bacterial key-taxa in stratified surface waters of the Southern Pacific Gyre.ISME J. 2024 Jan 8;18(1):wrae155. doi: 10.1093/ismejo/wrae155. ISME J. 2024. PMID: 39096506 Free PMC article.
-
Effects of Light and Dark Conditions on the Transcriptome of Aging Cultures of Candidatus Puniceispirillum marinum IMCC1322.J Microbiol. 2024 Apr;62(4):297-314. doi: 10.1007/s12275-024-00125-0. Epub 2024 Apr 25. J Microbiol. 2024. PMID: 38662311
-
Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota.ISME J. 2023 Jul;17(7):1063-1073. doi: 10.1038/s41396-023-01412-1. Epub 2023 Apr 29. ISME J. 2023. PMID: 37120702 Free PMC article.
-
Proteome Expression and Survival Strategies of a Proteorhodopsin-Containing Vibrio Strain under Carbon and Nitrogen Limitation.mSystems. 2022 Apr 26;7(2):e0126321. doi: 10.1128/msystems.01263-21. Epub 2022 Apr 6. mSystems. 2022. PMID: 35384695 Free PMC article.
-
Flavobacterium flabelliforme sp. nov. and Flavobacterium geliluteum sp. nov., Two Multidrug-Resistant Psychrotrophic Species Isolated From Antarctica.Front Microbiol. 2021 Oct 22;12:729977. doi: 10.3389/fmicb.2021.729977. eCollection 2021. Front Microbiol. 2021. PMID: 34745033 Free PMC article.
References
-
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. - PubMed
-
- Armstrong G.1999Carotenoid genetics and biochemistryIn: Cane DE (ed.).Isoprenoids Including Carotenoids and Steroids vol. 2Elsevier: Amsterdam, The Netherlands; 321–352.
-
- Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc Chem Res. 2002;35:113–120. - PubMed
-
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. - PubMed
-
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


