Mutant proteins as cancer-specific biomarkers
- PMID: 21248225
- PMCID: PMC3038743
- DOI: 10.1073/pnas.1019203108
Mutant proteins as cancer-specific biomarkers
Abstract
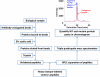
Cancer biomarkers are currently the subject of intense research because of their potential utility for diagnosis, prognosis, and targeted therapy. In theory, the gene products resulting from somatic mutations are the ultimate protein biomarkers, being not simply associated with tumors but actually responsible for tumorigenesis. We show here that the altered protein products resulting from somatic mutations can be identified directly and quantified by mass spectrometry. The peptides expressed from normal and mutant alleles were detected by selected reaction monitoring (SRM) of their product ions using a triple-quadrupole mass spectrometer. As a prototypical example of this approach, we demonstrated that it is possible to quantify the number and fraction of mutant Ras protein present in cancer cell lines. There were an average of 1.3 million molecules of Ras protein per cell, and the ratio of mutant to normal Ras proteins ranged from 0.49 to 5.6. Similarly, we found that mutant Ras proteins could be detected and quantified in clinical specimens such as colorectal and pancreatic tumor tissues as well as in premalignant pancreatic cyst fluids. In addition to answering basic questions about the relative levels of genetically abnormal proteins in tumors, this approach could prove useful for diagnostic applications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

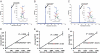

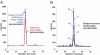
Similar articles
-
An improved quantitative mass spectrometry analysis of tumor specific mutant proteins at high sensitivity.Proteomics. 2012 May;12(9):1319-27. doi: 10.1002/pmic.201100611. Proteomics. 2012. PMID: 22589181
-
Targeted data-independent acquisition for mass spectrometric detection of RAS mutations in formalin-fixed, paraffin-embedded tumor biopsies.J Proteomics. 2018 Oct 30;189:91-96. doi: 10.1016/j.jprot.2018.04.022. Epub 2018 Apr 22. J Proteomics. 2018. PMID: 29684684
-
RAS signaling and anti-RAS therapy: lessons learned from genetically engineered mouse models, human cancer cells, and patient-related studies.Acta Biochim Biophys Sin (Shanghai). 2016 Jan;48(1):27-38. doi: 10.1093/abbs/gmv090. Epub 2015 Sep 7. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26350096 Free PMC article. Review.
-
GeLC-MRM quantitation of mutant KRAS oncoprotein in complex biological samples.J Proteome Res. 2012 Jul 6;11(7):3908-13. doi: 10.1021/pr300161j. Epub 2012 Jun 26. J Proteome Res. 2012. PMID: 22671702 Free PMC article.
-
RAS, wanted dead or alive: Advances in targeting RAS mutant cancers.Sci Signal. 2020 Mar 24;13(624):eaay6013. doi: 10.1126/scisignal.aay6013. Sci Signal. 2020. PMID: 32209699 Free PMC article. Review.
Cited by
-
Fast, sensitive detection of protein homologs using deep dense retrieval.Nat Biotechnol. 2024 Aug 9. doi: 10.1038/s41587-024-02353-6. Online ahead of print. Nat Biotechnol. 2024. PMID: 39123049
-
Detection and Quantitation of Endogenous Membrane-Bound RAS Proteins and KRAS Mutants in Cancer Cell Lines Using 1D-SDS-PAGE LC-MS2.Methods Mol Biol. 2024;2823:269-289. doi: 10.1007/978-1-0716-3922-1_17. Methods Mol Biol. 2024. PMID: 39052226
-
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.Sci Adv. 2024 Jul 12;10(28):eadk6580. doi: 10.1126/sciadv.adk6580. Epub 2024 Jul 10. Sci Adv. 2024. PMID: 38985864 Free PMC article.
-
Identification and verification of plasma protein biomarkers that accurately identify an ectopic pregnancy.Clin Proteomics. 2023 Sep 15;20(1):37. doi: 10.1186/s12014-023-09425-w. Clin Proteomics. 2023. PMID: 37715129 Free PMC article.
-
Dietary Oncopharmacognosy as a Crosswalk between Precision Oncology and Precision Nutrition.Nutrients. 2023 May 8;15(9):2219. doi: 10.3390/nu15092219. Nutrients. 2023. PMID: 37432381 Free PMC article. Review.
References
-
- Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA57345/CA/NCI NIH HHS/United States
- N01CN43302/CN/NCI NIH HHS/United States
- R01 CA130938/CA/NCI NIH HHS/United States
- U54 RR020839/RR/NCRR NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- CA62924/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 CA057345/CA/NCI NIH HHS/United States
- N01CN-43302/CN/NCI NIH HHS/United States
- CA130938/CA/NCI NIH HHS/United States
- R01 CA113669/CA/NCI NIH HHS/United States
- RR020839/RR/NCRR NIH HHS/United States
- R01 CA057345/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


