Pharmacogenomics: a systems approach
- PMID: 20836007
- PMCID: PMC3894835
- DOI: 10.1002/wsbm.42
Pharmacogenomics: a systems approach
Abstract
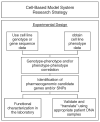
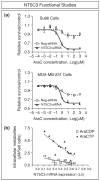
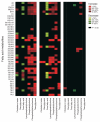
Pharmacogenetics and pharmacogenomics involve the study of the role of inheritance in individual variation in drug response, a phenotype that varies from potentially life-threatening adverse drug reactions to equally serious lack of therapeutic efficacy. Pharmacogenetics-pharmacogenomics represents a major component of the movement to 'individualized medicine'. Pharmacogenetic studies originally focused on monogenic traits, often involving genetic variation in drug metabolism. However, contemporary studies increasingly involve entire 'pathways' that include both pharmacokinetics (PKs)--factors that influence the concentration of a drug reaching its target(s)--and pharmacodynamics (PDs), factors associated with the drug target(s), as well as genome-wide approaches. The convergence of advances in pharmacogenetics with rapid developments in human genomics has resulted in the evolution of pharmacogenetics into pharmacogenomics. At the same time, studies of drug response are expanding beyond genomics to encompass pharmacotranscriptomics and pharmacometabolomics to become a systems-based discipline. This discipline is also increasingly moving across the 'translational interface' into the clinic and is being incorporated into the drug development process and governmental regulation of that process. The article will provide an overview of the development of pharmacogenetics-pharmacogenomics, the scientific advances that have contributed to the continuing evolution of this discipline, the incorporation of transcriptomic and metabolomic data into attempts to understand and predict variation in drug response phenotypes as well as challenges associated with the 'translation' of this important aspect of biomedical science into the clinic.
Figures

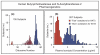
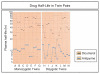
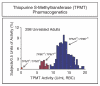
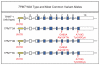

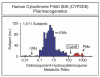

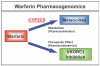
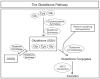
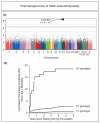
Similar articles
-
Pharmacogenetics and pharmacogenomics: development, science, and translation.Annu Rev Genomics Hum Genet. 2006;7:223-45. doi: 10.1146/annurev.genom.6.080604.162315. Annu Rev Genomics Hum Genet. 2006. PMID: 16948615 Review.
-
Pharmacogenomics: bench to bedside.Nat Rev Drug Discov. 2004 Sep;3(9):739-48. doi: 10.1038/nrd1497. Nat Rev Drug Discov. 2004. PMID: 15340384 Review.
-
Pharmacogenomics and individualized medicine: translating science into practice.Clin Pharmacol Ther. 2012 Oct;92(4):467-75. doi: 10.1038/clpt.2012.120. Epub 2012 Sep 5. Clin Pharmacol Ther. 2012. PMID: 22948889 Free PMC article.
-
Pharmacogenomics: catechol O-methyltransferase to thiopurine S-methyltransferase.Cell Mol Neurobiol. 2006 Jul-Aug;26(4-6):539-61. doi: 10.1007/s10571-006-9095-z. Epub 2006 Jun 29. Cell Mol Neurobiol. 2006. PMID: 16807786 Review.
-
Pharmacogenomics: candidate gene identification, functional validation and mechanisms.Hum Mol Genet. 2008 Oct 15;17(R2):R174-9. doi: 10.1093/hmg/ddn270. Hum Mol Genet. 2008. PMID: 18852207 Free PMC article. Review.
Cited by
-
Pharmacogenomics of Old and New Immunosuppressive Drugs for Precision Medicine in Kidney Transplantation.J Clin Med. 2023 Jul 3;12(13):4454. doi: 10.3390/jcm12134454. J Clin Med. 2023. PMID: 37445489 Free PMC article. Review.
-
COVID19 Drug Repository: text-mining the literature in search of putative COVID19 therapeutics.Nucleic Acids Res. 2021 Jan 8;49(D1):D1113-D1121. doi: 10.1093/nar/gkaa969. Nucleic Acids Res. 2021. PMID: 33166390 Free PMC article.
-
Application of Radiomics for Personalized Treatment of Cancer Patients.Cancer Manag Res. 2019 Dec 30;11:10851-10858. doi: 10.2147/CMAR.S232473. eCollection 2019. Cancer Manag Res. 2019. PMID: 31920394 Free PMC article. Review.
-
A Systems Biology Approach for Personalized Medicine in Refractory Epilepsy.Int J Mol Sci. 2019 Jul 30;20(15):3717. doi: 10.3390/ijms20153717. Int J Mol Sci. 2019. PMID: 31366017 Free PMC article. Review.
-
Use of Pharmacogenetic Drugs by the Dutch Population.Front Genet. 2019 Jul 2;10:567. doi: 10.3389/fgene.2019.00567. eCollection 2019. Front Genet. 2019. PMID: 31312209 Free PMC article.
References
-
- Guttmacher AE, Collins FS. Realising the promise of genomics in biomedical research. JAMA. 2005;294:1399–1402. - PubMed
-
- Weinshilboum RM. The therapeutic revolution. Clin Pharmacol Ther. 1987;42:481–484. - PubMed
-
- Vesell ES, Page JG. Genetic control of drug levels in man: antipyrine. Science. 1968;161:72–73. - PubMed
-
- Kalow W. Pharmacogenetics: Heredity and the Response to Drugs. WB Saunders; Philadelphia, London: 1962.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous


