Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain
- PMID: 20667828
- PMCID: PMC2951250
- DOI: 10.1074/jbc.M110.143313
Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain
Abstract
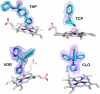
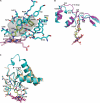
Cytochrome P450 46A1 (CYP46A1) initiates the major pathway of cholesterol elimination from the brain and thereby controls cholesterol turnover in this organ. We determined x-ray crystal structures of CYP46A1 in complex with four structurally distinct pharmaceuticals; antidepressant tranylcypromine (2.15 Å), anticonvulsant thioperamide (1.65 Å), antifungal voriconazole (2.35 Å), and antifungal clotrimazole (2.50 Å). All four drugs are nitrogen-containing compounds that have nanomolar affinity for CYP46A1 in vitro yet differ in size, shape, hydrophobicity, and type of the nitrogen ligand. Structures of the co-complexes demonstrate that each drug binds in a single orientation to the active site with tranylcypromine, thioperamide, and voriconazole coordinating the heme iron via their nitrogen atoms and clotrimazole being at a 4 Å distance from the heme iron. We show here that clotrimazole is also a substrate for CYP46A1. High affinity for CYP46A1 is determined by a set of specific interactions, some of which were further investigated by solution studies using structural analogs of the drugs and the T306A CYP46A1 mutant. Collectively, our results reveal how diverse inhibitors can be accommodated in the CYP46A1 active site and provide an explanation for the observed differences in the drug-induced spectral response. Co-complexes with tranylcypromine, thioperamide, and voriconazole represent the first structural characterization of the drug binding to a P450 enzyme.
Figures
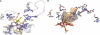
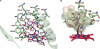
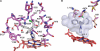
Similar articles
-
Antifungal Azoles: Structural Insights into Undesired Tight Binding to Cholesterol-Metabolizing CYP46A1.Mol Pharmacol. 2013 Jul;84(1):86-94. doi: 10.1124/mol.113.085902. Epub 2013 Apr 19. Mol Pharmacol. 2013. PMID: 23604141 Free PMC article.
-
In silico and intuitive predictions of CYP46A1 inhibition by marketed drugs with subsequent enzyme crystallization in complex with fluvoxamine.Mol Pharmacol. 2012 Nov;82(5):824-34. doi: 10.1124/mol.112.080424. Epub 2012 Aug 2. Mol Pharmacol. 2012. PMID: 22859721 Free PMC article.
-
The antifungal drug voriconazole is an efficient inhibitor of brain cholesterol 24S-hydroxylase in vitro and in vivo.J Lipid Res. 2010 Feb;51(2):318-23. doi: 10.1194/jlr.M900174-JLR200. Epub 2009 May 27. J Lipid Res. 2010. PMID: 19474457 Free PMC article.
-
Cholesterol 24-Hydroxylation by CYP46A1: Benefits of Modulation for Brain Diseases.Neurotherapeutics. 2019 Jul;16(3):635-648. doi: 10.1007/s13311-019-00731-6. Neurotherapeutics. 2019. PMID: 31001737 Free PMC article. Review.
-
Cholesterol-metabolizing cytochromes P450.Drug Metab Dispos. 2006 Apr;34(4):513-20. doi: 10.1124/dmd.105.008789. Epub 2006 Jan 24. Drug Metab Dispos. 2006. PMID: 16434543 Review.
Cited by
-
Identification of potential inhibitors of brain-specific CYP46A1 from phytoconstituents in Indian traditional medicinal plants.J Proteins Proteom. 2022;13(4):227-245. doi: 10.1007/s42485-022-00098-x. Epub 2022 Nov 16. J Proteins Proteom. 2022. PMID: 36404953 Free PMC article.
-
Assessment of cholesterol homeostasis in the living human brain.Sci Transl Med. 2022 Oct 5;14(665):eadc9967. doi: 10.1126/scitranslmed.adc9967. Epub 2022 Oct 5. Sci Transl Med. 2022. PMID: 36197966 Free PMC article.
-
Remembering your A, B, C's: Alzheimer's disease and ABCA1.Acta Pharm Sin B. 2022 Mar;12(3):995-1018. doi: 10.1016/j.apsb.2022.01.011. Epub 2022 Jan 24. Acta Pharm Sin B. 2022. PMID: 35530134 Free PMC article. Review.
-
The Hydroxylation Position Rather than Chirality Determines How Efavirenz Metabolites Activate Cytochrome P450 46A1 In Vitro.Drug Metab Dispos. 2022 Jul;50(7):923-930. doi: 10.1124/dmd.122.000874. Epub 2022 Apr 29. Drug Metab Dispos. 2022. PMID: 35489779 Free PMC article.
-
Cholesterol Hydroxylating Cytochrome P450 46A1: From Mechanisms of Action to Clinical Applications.Front Aging Neurosci. 2021 Jul 8;13:696778. doi: 10.3389/fnagi.2021.696778. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34305573 Free PMC article. Review.
References
-
- Björkhem I., Lütjohann D., Diczfalusy U., Ståhle L., Ahlborg G., Wahren J. (1998) J. Lipid Res. 39, 1594–1600 - PubMed
-
- Meaney S., Bodin K., Diczfalusy U., Björkhem I. (2002) J. Lipid Res. 43, 2130–2135 - PubMed
-
- Xie C., Lund E. G., Turley S. D., Russell D. W., Dietschy J. M. (2003) J. Lipid Res. 44, 1780–1789 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases


