Distribution of fitness effects caused by single-nucleotide substitutions in bacteriophage f1
- PMID: 20382832
- PMCID: PMC2881140
- DOI: 10.1534/genetics.110.115162
Distribution of fitness effects caused by single-nucleotide substitutions in bacteriophage f1
Abstract
Empirical knowledge of the fitness effects of mutations is important for understanding many evolutionary processes, yet this knowledge is often hampered by several sources of measurement error and bias. Most of these problems can be solved using site-directed mutagenesis to engineer single mutations, an approach particularly suited for viruses due to their small genomes. Here, we used this technique to measure the fitness effect of 100 single-nucleotide substitutions in the bacteriophage f1, a filamentous single-strand DNA virus. We found that approximately one-fifth of all mutations are lethal. Viable ones reduced fitness by 11% on average and were accurately described by a log-normal distribution. More than 90% of synonymous substitutions were selectively neutral, while those affecting intergenic regions reduced fitness by 14% on average. Mutations leading to amino acid substitutions had an overall mean deleterious effect of 37%, which increased to 45% for those changing the amino acid polarity. Interestingly, mutations affecting early steps of the infection cycle tended to be more deleterious than those affecting late steps. Finally, we observed at least two beneficial mutations. Our results confirm that high mutational sensitivity is a general property of viruses with small genomes, including RNA and single-strand DNA viruses infecting animals, plants, and bacteria.
Figures
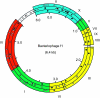
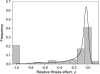
Similar articles
-
The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages.PLoS Genet. 2009 Nov;5(11):e1000742. doi: 10.1371/journal.pgen.1000742. Epub 2009 Nov 26. PLoS Genet. 2009. PMID: 19956760 Free PMC article.
-
Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies.Philos Trans R Soc Lond B Biol Sci. 2010 Jun 27;365(1548):1975-82. doi: 10.1098/rstb.2010.0063. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20478892 Free PMC article. Review.
-
The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8396-401. doi: 10.1073/pnas.0400146101. Epub 2004 May 24. Proc Natl Acad Sci U S A. 2004. PMID: 15159545 Free PMC article.
-
The fitness effects of synonymous mutations in DNA and RNA viruses.Mol Biol Evol. 2012 Jan;29(1):17-20. doi: 10.1093/molbev/msr179. Epub 2011 Jul 18. Mol Biol Evol. 2012. PMID: 21771719
-
The distribution of fitness effects of new mutations.Nat Rev Genet. 2007 Aug;8(8):610-8. doi: 10.1038/nrg2146. Nat Rev Genet. 2007. PMID: 17637733 Review.
Cited by
-
Fit-Seq2.0: An Improved Software for High-Throughput Fitness Measurements Using Pooled Competition Assays.J Mol Evol. 2023 Jun;91(3):334-344. doi: 10.1007/s00239-023-10098-0. Epub 2023 Mar 6. J Mol Evol. 2023. PMID: 36877292 Free PMC article.
-
Fitness and Functional Landscapes of the E. coli RNase III Gene rnc.Mol Biol Evol. 2023 Mar 4;40(3):msad047. doi: 10.1093/molbev/msad047. Mol Biol Evol. 2023. PMID: 36848192 Free PMC article.
-
COVID-19: A state of art on immunological responses, mutations, and treatment modalities in riposte.J Infect Public Health. 2023 Feb;16(2):233-249. doi: 10.1016/j.jiph.2022.12.019. Epub 2022 Dec 29. J Infect Public Health. 2023. PMID: 36603376 Free PMC article. Review.
-
The High Mutational Sensitivity of ccdA Antitoxin Is Linked to Codon Optimality.Mol Biol Evol. 2022 Oct 7;39(10):msac187. doi: 10.1093/molbev/msac187. Mol Biol Evol. 2022. PMID: 36069948 Free PMC article.
-
A White Plaque, Associated with Genomic Deletion, Derived from M13KE-Based Peptide Library Is Enriched in a Target-Unrelated Manner during Phage Display Biopanning Due to Propagation Advantage.Int J Mol Sci. 2022 Mar 18;23(6):3308. doi: 10.3390/ijms23063308. Int J Mol Sci. 2022. PMID: 35328728 Free PMC article.
References
-
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57 289–300.
-
- Bull, J. J., M. R. Badgett, R. Springman and I. J. Molineux, 2004. Genome properties and the limits of adaptation in bacteriophages. Evolution 58 692–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources


