Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea
- PMID: 20305030
- PMCID: PMC2869143
- DOI: 10.1128/AEM.02971-09
Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea
Abstract
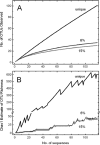
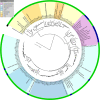
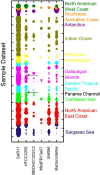
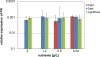
Proteorhodopsin (PR), a photoactive proton pump containing retinal, is present in approximately half of all bacteria in the ocean, but its physiological role is still unclear, since very few strains carrying the PR gene have been cultured. The aim of this work was to characterize PR diversity in a North Sea water sample, cultivate a strain representative of North Sea PR clusters, and study the effects of light and carbon concentration on the expression of the PR gene. A total of 117 PR sequences, of which 101 were unique, were obtained from a clone library of PCR-amplified PR gene fragments. Of the North Sea PRs, 97% were green light absorbing, as inferred from the amino acid at position 105; 67% of the PR protein fragments showed closest similarity to PRs from Alphaproteobacteria, 4% showed closest similarity to PRs from Gammaproteobacteria, and 29% showed closest similarity to PRs from "Bacteroidetes"/Flavobacteria. The dominant PR cluster (comprising 18% of all PRs) showed a high degree of similarity to the PR from the cultivated Roseobacter strain HTCC2255. The relative abundances of the North Sea PR clusters were confirmed by quantitative PCR. They were detected in metagenomic fragments from coastal oceans worldwide with various degrees of abundance. Several hundred bacterial strains from the North Sea water sample were cultivated on oligocarbophilic media. By screening with degenerate primers, two strains carrying the PR gene were identified. Their 16S rRNA gene sequences were identical and affiliated with a Bacteroidetes subcluster from the North Sea. The PR sequence of isolate PRO95 was completed by chromosomal walking. It was 76% identical to that of Dokdonia donghaensis MED134 and was functional, as indicated by the signature amino acids. PRO95 expressed its PR gene in liquid media containing between 9.7 and 121 mM carbon, both in the light and in the dark. Growth was not enhanced by light. Thus, the detection of the physiological role of PR may require more sensitive methods.
Figures


Similar articles
-
Abundant proteorhodopsin genes in the North Atlantic Ocean.Environ Microbiol. 2008 Jan;10(1):99-109. doi: 10.1111/j.1462-2920.2007.01436.x. Environ Microbiol. 2008. PMID: 18211270
-
Diversity and functional analysis of proteorhodopsin in marine Flavobacteria.Environ Microbiol. 2012 May;14(5):1240-8. doi: 10.1111/j.1462-2920.2012.02702.x. Epub 2012 Feb 13. Environ Microbiol. 2012. PMID: 22329552
-
Light stimulates growth of proteorhodopsin-containing marine Flavobacteria.Nature. 2007 Jan 11;445(7124):210-3. doi: 10.1038/nature05381. Nature. 2007. PMID: 17215843
-
Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology.Microbiol Mol Biol Rev. 2016 Sep 14;80(4):929-54. doi: 10.1128/MMBR.00003-16. Print 2016 Dec. Microbiol Mol Biol Rev. 2016. PMID: 27630250 Free PMC article. Review.
-
Evidence for the ubiquity of mixotrophic bacteria in the upper ocean: implications and consequences.Appl Environ Microbiol. 2006 Dec;72(12):7431-7. doi: 10.1128/AEM.01559-06. Epub 2006 Oct 6. Appl Environ Microbiol. 2006. PMID: 17028233 Free PMC article. Review. No abstract available.
Cited by
-
Effects of Light and Dark Conditions on the Transcriptome of Aging Cultures of Candidatus Puniceispirillum marinum IMCC1322.J Microbiol. 2024 Apr;62(4):297-314. doi: 10.1007/s12275-024-00125-0. Epub 2024 Apr 25. J Microbiol. 2024. PMID: 38662311
-
Scramblase activity of proteorhodopsin confers physiological advantages to Escherichia coli in the absence of light.iScience. 2023 Nov 22;26(12):108551. doi: 10.1016/j.isci.2023.108551. eCollection 2023 Dec 15. iScience. 2023. PMID: 38125024 Free PMC article.
-
Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota.ISME J. 2023 Jul;17(7):1063-1073. doi: 10.1038/s41396-023-01412-1. Epub 2023 Apr 29. ISME J. 2023. PMID: 37120702 Free PMC article.
-
Ecological divergence of syntopic marine bacterial species is shaped by gene content and expression.ISME J. 2023 Jun;17(6):813-822. doi: 10.1038/s41396-023-01390-4. Epub 2023 Mar 4. ISME J. 2023. PMID: 36871069 Free PMC article.
-
Proteorhodopsin Phototrophy in Antarctic Coastal Waters.mSphere. 2021 Aug 25;6(4):e0052521. doi: 10.1128/mSphere.00525-21. Epub 2021 Aug 18. mSphere. 2021. PMID: 34406852 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Atamna-Ismaeel, N., G. Sabehi, I. Sharon, K. P. Witzel, M. Labrenz, K. Jurgens, T. Barkay, M. Stomp, J. Huisman, and O. Beja. 2008. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2:656-662. - PubMed
-
- Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. - PubMed
-
- Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials


