MicroRNAs: tools for cancer diagnostics
- PMID: 19834118
- PMCID: PMC2802653
- DOI: 10.1136/gut.2009.179531
MicroRNAs: tools for cancer diagnostics
Abstract
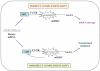
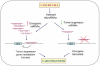
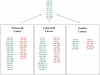
Recently, a novel class of global gene regulators called microRNAs (miRNAs), were identified in both plants and animals. MiRNAs can reduce protein levels of their target genes with a minor impact on the target genes' mRNAs. Accumulating evidence demonstrates the importance of miRNAs in cancer. MiRNAs that are overexpressed in cancer may function as oncogenes, and miRNAs with tumour suppressor activity in normal tissue may be downregulated in cancer. Although major advances have been achieved in our understanding of cancer biology, as well as in the development of new targeted therapies, the progress in developing improved early diagnosis and screening tests has been inadequate. This results in most cancers being diagnosed in advanced stages, delaying timely treatment and leading to poor outcomes. There is intense research seeking specific molecular changes that are able to identify patients with early cancer or precursor lesions. MiRNA expression data in various cancers demonstrate that cancer cells have different miRNA profiles compared with normal cells, thus underscoring the tremendous diagnostic and therapeutic potential of miRNAs in cancer. These unique properties of miRNAs make them extremely useful potential agents for clinical diagnostics as well as in personalised care for individual patients in the future.
Figures


Similar articles
-
Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of lung cancer.Oncol Rep. 2017 Jun;37(6):3543-3553. doi: 10.3892/or.2017.5612. Epub 2017 Apr 28. Oncol Rep. 2017. PMID: 28498428
-
Global Analysis of miRNA-mRNA Interaction Network in Breast Cancer with Brain Metastasis.Anticancer Res. 2017 Aug;37(8):4455-4468. doi: 10.21873/anticanres.11841. Anticancer Res. 2017. PMID: 28739740
-
miRNA expression profile changes in the peripheral blood of monozygotic discordant twins for epithelial ovarian carcinoma: potential new biomarkers for early diagnosis and prognosis of ovarian carcinoma.J Ovarian Res. 2020 Aug 27;13(1):99. doi: 10.1186/s13048-020-00706-8. J Ovarian Res. 2020. PMID: 32854743 Free PMC article.
-
MicroRNAs and cancer; an overview.Curr Pharm Biotechnol. 2014;15(5):430-7. doi: 10.2174/1389201015666140519095304. Curr Pharm Biotechnol. 2014. PMID: 24846068 Review.
-
microRNAs as oncogenes and tumor suppressors.Dev Biol. 2007 Feb 1;302(1):1-12. doi: 10.1016/j.ydbio.2006.08.028. Epub 2006 Aug 16. Dev Biol. 2007. PMID: 16989803 Review.
Cited by
-
Paper-Based DNA Biosensor for Rapid and Selective Detection of miR-21.Biosensors (Basel). 2024 Oct 8;14(10):485. doi: 10.3390/bios14100485. Biosensors (Basel). 2024. PMID: 39451697 Free PMC article.
-
Evaluation of the Rock1 and microRNA-148a expression in biopsies collected from patients with Helicobacter pylori induced gastritis.BMC Gastroenterol. 2024 Aug 7;24(1):251. doi: 10.1186/s12876-024-03347-z. BMC Gastroenterol. 2024. PMID: 39112943 Free PMC article.
-
Engineering a cell-free biosensor signal amplification circuit with polymerase strand recycling.bioRxiv [Preprint]. 2024 Apr 25:2024.04.25.591074. doi: 10.1101/2024.04.25.591074. bioRxiv. 2024. PMID: 38712145 Free PMC article. Preprint.
-
microRNA-184 in the landscape of human malignancies: a review to roles and clinical significance.Cell Death Discov. 2023 Nov 24;9(1):423. doi: 10.1038/s41420-023-01718-1. Cell Death Discov. 2023. PMID: 38001121 Free PMC article. Review.
-
NF-κB downstream miR-1262 disturbs colon cancer cell malignant behaviors by targeting FGFR1.Acta Biochim Biophys Sin (Shanghai). 2023 Nov 25;55(11):1819-1832. doi: 10.3724/abbs.2023235. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37867436 Free PMC article.
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed
-
- Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. - PubMed
-
- Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. - PubMed
-
- Berezikov E, Guryev V, van de Belt J, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

