Whirly proteins maintain plastid genome stability in Arabidopsis
- PMID: 19666500
- PMCID: PMC2732811
- DOI: 10.1073/pnas.0901710106
Whirly proteins maintain plastid genome stability in Arabidopsis
Abstract
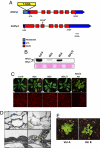
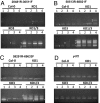
Maintenance of genome stability is essential for the accurate propagation of genetic information and cell growth and survival. Organisms have therefore developed efficient strategies to prevent DNA lesions and rearrangements. Much of the information concerning these strategies has been obtained through the study of bacterial and nuclear genomes. Comparatively, little is known about how organelle genomes maintain a stable structure. Here, we report that the plastid-localized Whirly ssDNA-binding proteins are required for plastid genome stability in Arabidopsis. We show that a double KO of the genes AtWhy1 and AtWhy3 leads to the appearance of plants with variegated green/white/yellow leaves, symptomatic of nonfunctional chloroplasts. This variegation is maternally inherited, indicating defects in the plastid genome. Indeed, in all variegated lines examined, reorganized regions of plastid DNA are amplified as circular and/or head-tail concatemers. All amplified regions are delimited by short direct repeats of 10-18 bp, strongly suggesting that these regions result from illegitimate recombination between repeated sequences. This type of recombination occurs frequently in plants lacking both Whirlies, to a lesser extent in single KO plants and rarely in WT individuals. Maize mutants for the ZmWhy1 Whirly protein also show an increase in the frequency of illegitimate recombination. We propose a model where Whirlies contribute to plastid genome stability by protecting against illegitimate repeat-mediated recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis.Nucleic Acids Res. 2008 Sep;36(16):5152-65. doi: 10.1093/nar/gkn492. Epub 2008 Aug 2. Nucleic Acids Res. 2008. PMID: 18676978 Free PMC article.
-
Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns.Plant Physiol. 2006 Dec;142(4):1656-63. doi: 10.1104/pp.106.088096. Epub 2006 Oct 27. Plant Physiol. 2006. PMID: 17071648 Free PMC article.
-
Genes Sufficient for Synthesizing Peptidoglycan are Retained in Gymnosperm Genomes, and MurE from Larix gmelinii can Rescue the Albino Phenotype of Arabidopsis MurE Mutation.Plant Cell Physiol. 2017 Mar 1;58(3):587-597. doi: 10.1093/pcp/pcx005. Plant Cell Physiol. 2017. PMID: 28158764
-
Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana.Genes Genet Syst. 2003 Feb;78(1):1-9. doi: 10.1266/ggs.78.1. Genes Genet Syst. 2003. PMID: 12655133 Review.
-
PPR-SMRs: ancient proteins with enigmatic functions.RNA Biol. 2013;10(9):1501-10. doi: 10.4161/rna.26172. Epub 2013 Aug 28. RNA Biol. 2013. PMID: 24004908 Free PMC article. Review.
Cited by
-
Complete chloroplast genomes of three Polygala species and indel marker development for identification of authentic polygalae radix (Polygala tenuifolia).Genes Genomics. 2024 Nov 14. doi: 10.1007/s13258-024-01573-z. Online ahead of print. Genes Genomics. 2024. PMID: 39543069
-
Genome-Wide Identification and Molecular Evolutionary History of the Whirly Family Genes in Brassica napus.Plants (Basel). 2024 Aug 13;13(16):2243. doi: 10.3390/plants13162243. Plants (Basel). 2024. PMID: 39204679 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the Sweet Cherry Whirly Gene Family.Curr Issues Mol Biol. 2024 Jul 26;46(8):8015-8030. doi: 10.3390/cimb46080474. Curr Issues Mol Biol. 2024. PMID: 39194691 Free PMC article.
-
Genome-Wide Identification of the Whirly Gene Family and Its Potential Function in Low Phosphate Stress in Soybean (Glycine max).Genes (Basel). 2024 Jun 25;15(7):833. doi: 10.3390/genes15070833. Genes (Basel). 2024. PMID: 39062612 Free PMC article.
-
Evolution of Whirly1 in the angiosperms: sequence, splicing, and expression in a clade of early transitional mycoheterotrophic orchids.Front Plant Sci. 2024 Jun 28;15:1241515. doi: 10.3389/fpls.2024.1241515. eCollection 2024. Front Plant Sci. 2024. PMID: 39006962 Free PMC article.
References
-
- Day A, Madesis P. DNA replication, recombination, and repair in plastids. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol 19. Berlin: Springer; 2007. pp. 65–119.
-
- Palmer JD. Chloroplast DNA exists in 2 orientations. Nature. 1983;301:92–93.
-
- Cao J, Combs C, Jagendorf AT. The chloroplast-located homolog of bacterial DNA recombinase. Plant Cell Physiol. 1997;38:1319–1325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials


