Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice
- PMID: 19494335
- PMCID: PMC2792900
- DOI: 10.4049/jimmunol.0900263
Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice
Abstract
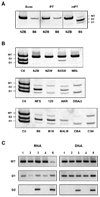
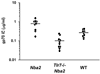
Endogenous retroviruses are implicated in the pathogenesis of systemic lupus erythematosus (SLE). Because four different classes of endogenous retroviruses, i.e., ecotropic, xenotropic, polytropic, or modified polytropic (mPT), are expressed in mice, we investigated the possibility that a particular class of endogenous retroviruses is associated with the development of murine SLE. We observed >15-fold increased expression of mPT env (envelope) RNA in livers of all four lupus-prone mice, as compared with those of nine nonautoimmune strains of mice. This was not the case for the three other classes of retroviruses. Furthermore, we found that in addition to intact mPT transcripts, many strains of mice expressed two defective mPT env transcripts which carry a deletion in the env sequence of the 3' portion of the gp70 surface protein and the 5' portion of the p15E transmembrane protein, respectively. Remarkably, in contrast to nonautoimmune strains of mice, all four lupus-prone mice expressed abundant levels of intact mPT env transcripts, but only low or nondetectable levels of the mutant env transcripts. The Sgp3 (serum gp70 production 3) locus derived from lupus-prone mice was responsible for the selective up-regulation of the intact mPT env RNA. Finally, we observed that single-stranded RNA-specific TLR7 played a critical role in the production of anti-gp70 autoantibodies. These data suggest that lupus-prone mice may possess a unique genetic mechanism responsible for the expression of mPT retroviruses, which could act as a triggering factor through activating TLR7 for the development of autoimmune responses in mice predisposed to SLE.
Figures





Similar articles
-
Sgp3 and TLR7 stimulation differentially alter the expression profile of modified polytropic retroviruses implicated in murine systemic lupus.J Autoimmun. 2012 Jun;38(4):361-8. doi: 10.1016/j.jaut.2012.03.002. Epub 2012 Apr 13. J Autoimmun. 2012. PMID: 22503566 Free PMC article.
-
Three Sgp loci act independently as well as synergistically to elevate the expression of specific endogenous retroviruses implicated in murine lupus.J Autoimmun. 2013 Jun;43:10-7. doi: 10.1016/j.jaut.2013.01.014. Epub 2013 Mar 7. J Autoimmun. 2013. PMID: 23465716 Free PMC article.
-
TLR-mediated up-regulation of serum retroviral gp70 is controlled by the Sgp loci of lupus-prone mice.J Autoimmun. 2010 Sep;35(2):153-9. doi: 10.1016/j.jaut.2010.06.001. Epub 2010 Jul 8. J Autoimmun. 2010. PMID: 20619604 Free PMC article.
-
Role of endogenous retroviruses in murine SLE.Autoimmun Rev. 2010 Nov;10(1):27-34. doi: 10.1016/j.autrev.2010.07.012. Epub 2010 Jul 24. Autoimmun Rev. 2010. PMID: 20659589 Review.
-
Emerging roles of TLR7 and TLR9 in murine SLE.J Autoimmun. 2009 Nov-Dec;33(3-4):231-8. doi: 10.1016/j.jaut.2009.10.001. Epub 2009 Oct 21. J Autoimmun. 2009. PMID: 19846276 Review.
Cited by
-
Reactivated endogenous retroviruses promote protein aggregate spreading.Nat Commun. 2023 Aug 18;14(1):5034. doi: 10.1038/s41467-023-40632-z. Nat Commun. 2023. PMID: 37596282 Free PMC article.
-
Human endogenous retroviruses and the inflammatory response: A vicious circle associated with health and illness.Front Immunol. 2022 Nov 23;13:1057791. doi: 10.3389/fimmu.2022.1057791. eCollection 2022. Front Immunol. 2022. PMID: 36518758 Free PMC article. Review.
-
Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability.Nat Cell Biol. 2022 Jan;24(1):99-111. doi: 10.1038/s41556-021-00795-7. Epub 2021 Dec 27. Nat Cell Biol. 2022. PMID: 34961794 Free PMC article.
-
T Cell Homeostatic Proliferation Promotes a Redox State That Drives Metabolic and Epigenetic Upregulation of Inflammatory Pathways in Lupus.Antioxid Redox Signal. 2022 Mar;36(7-9):410-422. doi: 10.1089/ars.2021.0078. Epub 2021 Nov 9. Antioxid Redox Signal. 2022. PMID: 34328790 Free PMC article. Review.
-
Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota.Cell. 2021 Jul 8;184(14):3794-3811.e19. doi: 10.1016/j.cell.2021.05.020. Epub 2021 Jun 23. Cell. 2021. PMID: 34166614 Free PMC article.
References
-
- Maruyama N, Furukawa F, Nakai Y, Sasaki Y, Ohta K, Ozaki S, Hirose S, Shirai T. Genetic studies of autoimmunity in New Zealand mice. IV. Contribution of NZB and NZW genes to the spontaneous occurrence of retroviral gp70 immune complexes in (NZB × NZW)F1 hybrid and the correlation to renal disease. J. Immunol. 1983;130:740–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases


