Regulation of translation initiation by RNA binding proteins
- PMID: 19385727
- PMCID: PMC4682898
- DOI: 10.1146/annurev.micro.091208.073514
Regulation of translation initiation by RNA binding proteins
Abstract
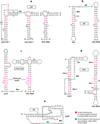
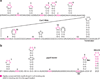
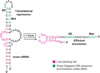
RNA binding proteins are capable of regulating translation initiation by a variety of mechanisms. Although the vast majority of these regulatory mechanisms involve translational repression, one example of translational activation has been characterized in detail. The RNA recognition targets of these regulatory proteins exhibit a wide range in structural complexity, with some proteins recognizing complex pseudoknot structures and others binding to simple RNA hairpins and/or short repeated single-stranded sequences. In some instances the bound protein directly competes with ribosome binding, and in other instances the bound protein promotes formation of an RNA structure that inhibits ribosome binding. Examples also exist in which the bound protein traps the ribosome in a complex that is incapable of initiating translation.
Figures



Similar articles
-
Construction of regulatable picornavirus IRESes as a test of current models of the mechanism of internal translation initiation.RNA. 2001 May;7(5):647-60. doi: 10.1017/s1355838201001911. RNA. 2001. PMID: 11350029 Free PMC article.
-
Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo.RNA. 2000 Oct;6(10):1380-92. doi: 10.1017/s1355838200000753. RNA. 2000. PMID: 11073214 Free PMC article.
-
Gene regulation: translational initiation by internal ribosome binding.Curr Opin Genet Dev. 1993 Apr;3(2):295-300. doi: 10.1016/0959-437x(93)90037-p. Curr Opin Genet Dev. 1993. PMID: 8504255 Free PMC article. Review.
-
Structured mRNAs regulate translation initiation by binding to the platform of the ribosome.Cell. 2007 Sep 21;130(6):1019-31. doi: 10.1016/j.cell.2007.07.008. Cell. 2007. PMID: 17889647
-
From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements.Crit Rev Biochem Mol Biol. 2006 Nov-Dec;41(6):329-38. doi: 10.1080/10409230600914294. Crit Rev Biochem Mol Biol. 2006. PMID: 17092822 Review.
Cited by
-
Stress-Induced Eukaryotic Translational Regulatory Mechanisms.J Clin Med Sci. 2024;8(2):1000277. Epub 2024 Jun 24. J Clin Med Sci. 2024. PMID: 39364184 Free PMC article.
-
Role of Small Non-Coding RNA in Gram-Negative Bacteria: New Insights and Comprehensive Review of Mechanisms, Functions, and Potential Applications.Mol Biotechnol. 2024 Aug 17. doi: 10.1007/s12033-024-01248-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39153013 Review.
-
RNA binding proteins as mediators of pathological cardiac remodeling.Front Cell Dev Biol. 2024 May 16;12:1368097. doi: 10.3389/fcell.2024.1368097. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38818408 Free PMC article. Review.
-
Stress-induced Eukaryotic Translational Regulatory Mechanisms.ArXiv [Preprint]. 2024 May 2:arXiv:2405.01664v1. ArXiv. 2024. Update in: J Clin Med Sci. 2024;8(2):1000277.. PMID: 38745702 Free PMC article. Updated. Preprint.
-
RNA cis-regulators are important for Streptococcus pneumoniae in vivo success.PLoS Genet. 2024 Mar 5;20(3):e1011188. doi: 10.1371/journal.pgen.1011188. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38442125 Free PMC article.
References
-
- Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. - PubMed
-
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


