Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350
- PMID: 18375553
- PMCID: PMC2395044
- DOI: 10.1128/JB.00204-08
Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350
Abstract
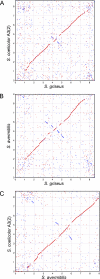
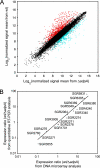
We determined the complete genome sequence of Streptomyces griseus IFO 13350, a soil bacterium producing an antituberculosis agent, streptomycin, which is the first aminoglycoside antibiotic, discovered more than 60 years ago. The linear chromosome consists of 8,545,929 base pairs (bp), with an average G+C content of 72.2%, predicting 7,138 open reading frames, six rRNA operons (16S-23S-5S), and 66 tRNA genes. It contains extremely long terminal inverted repeats (TIRs) of 132,910 bp each. The telomere's nucleotide sequence and secondary structure, consisting of several palindromes with a loop sequence of 5'-GGA-3', are different from those of typical telomeres conserved among other Streptomyces species. In accordance with the difference, the chromosome has pseudogenes for a conserved terminal protein (Tpg) and a telomere-associated protein (Tap), and a novel pair of Tpg and Tap proteins is instead encoded by the TIRs. Comparisons with the genomes of two related species, Streptomyces coelicolor A3(2) and Streptomyces avermitilis, clarified not only the characteristics of the S. griseus genome but also the existence of 24 Streptomyces-specific proteins. The S. griseus genome contains 34 gene clusters or genes for the biosynthesis of known or unknown secondary metabolites. Transcriptome analysis using a DNA microarray showed that at least four of these clusters, in addition to the streptomycin biosynthesis gene cluster, were activated directly or indirectly by AdpA, which is a central transcriptional activator for secondary metabolism and morphogenesis in the A-factor (a gamma-butyrolactone signaling molecule) regulatory cascade in S. griseus.
Figures

Similar articles
-
A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus.Front Biosci. 2002 Oct 1;7:d2045-57. doi: 10.2741/A897. Front Biosci. 2002. PMID: 12165483 Review.
-
Streptomycin production by Streptomyces griseus can be modulated by a mechanism not associated with change in the adpA component of the A-factor cascade.Biotechnol Lett. 2007 Jan;29(1):57-64. doi: 10.1007/s10529-006-9216-2. Epub 2006 Nov 22. Biotechnol Lett. 2007. PMID: 17120093
-
The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus : identification of a target gene of the A-factor receptor.Mol Microbiol. 1999 Oct;34(1):102-11. doi: 10.1046/j.1365-2958.1999.01579.x. Mol Microbiol. 1999. PMID: 10540289
-
Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus.J Bacteriol. 2005 Aug;187(16):5595-604. doi: 10.1128/JB.187.16.5595-5604.2005. J Bacteriol. 2005. PMID: 16077104 Free PMC article.
-
A-factor and streptomycin biosynthesis in Streptomyces griseus.Antonie Van Leeuwenhoek. 1993-1994;64(2):177-86. doi: 10.1007/BF00873026. Antonie Van Leeuwenhoek. 1993. PMID: 8092858 Review.
Cited by
-
Activation of Secondary Metabolism and Protease Activity Mechanisms in the Black Koji Mold Aspergillus luchuensis through Coculture with Animal Cells.ACS Omega. 2024 Oct 10;9(42):43129-43137. doi: 10.1021/acsomega.4c07124. eCollection 2024 Oct 22. ACS Omega. 2024. PMID: 39464474 Free PMC article.
-
Streptomyces sp. from desert soil as a biofactory for antioxidants with radical scavenging and iron chelating potential.BMC Microbiol. 2024 Oct 21;24(1):419. doi: 10.1186/s12866-024-03586-w. BMC Microbiol. 2024. PMID: 39434054 Free PMC article.
-
Discovery of a novel methionine biosynthetic route via O-phospho-l-homoserine.Appl Environ Microbiol. 2024 Oct 23;90(10):e0124724. doi: 10.1128/aem.01247-24. Epub 2024 Sep 23. Appl Environ Microbiol. 2024. PMID: 39311576
-
Potential of Streptomyces avermitilis: A Review on Avermectin Production and Its Biocidal Effect.Metabolites. 2024 Jun 30;14(7):374. doi: 10.3390/metabo14070374. Metabolites. 2024. PMID: 39057697 Free PMC article. Review.
-
Sequence and origin of the Streptomyces intergenetic-conjugation helper plasmid pUZ8002.Access Microbiol. 2024 Jun 5;6(6):000808.v3. doi: 10.1099/acmi.0.000808.v3. eCollection 2024. Access Microbiol. 2024. PMID: 39045241 Free PMC article.
References
-
- Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. - PubMed
-
- Brüggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Dürre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305671-673. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous


