Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria
- PMID: 18348319
- PMCID: PMC2799225
- DOI: 10.1002/pmic.200700851
Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria
Abstract
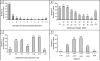
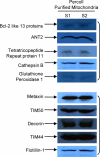
Mitochondria play essential roles in cardiac pathophysiology and the murine model has been extensively used to investigate cardiovascular diseases. In the present study, we characterized murine cardiac mitochondria using an LC/MS/MS approach. We extracted and purified cardiac mitochondria; validated their functionality to ensure the final preparation contains necessary components to sustain their normal function; and subjected these validated organelles to LC/MS/MS-based protein identification. A total of 940 distinct proteins were identified from murine cardiac mitochondria, among which, 480 proteins were not previously identified by major proteomic profiling studies. The 940 proteins consist of functional clusters known to support oxidative phosphorylation, metabolism, and biogenesis. In addition, there are several other clusters, including proteolysis, protein folding, and reduction/oxidation signaling, which ostensibly represent previously under-appreciated tasks of cardiac mitochondria. Moreover, many identified proteins were found to occupy other subcellular locations, including cytoplasm, ER, and golgi, in addition to their presence in the mitochondria. These results provide a comprehensive picture of the murine cardiac mitochondrial proteome and underscore tissue- and species-specification. Moreover, the use of functionally intact mitochondria insures that the proteomic observations in this organelle are relevant to its normal biology and facilitates decoding the interplay between mitochondria and other organelles.
Figures



Similar articles
-
Altered proteome biology of cardiac mitochondria under stress conditions.J Proteome Res. 2008 Jun;7(6):2204-14. doi: 10.1021/pr070371f. Epub 2008 May 17. J Proteome Res. 2008. PMID: 18484766 Free PMC article.
-
Proteomic analysis of mitochondria from Caenorhabditis elegans.Proteomics. 2009 Oct;9(19):4539-53. doi: 10.1002/pmic.200900101. Proteomics. 2009. PMID: 19670372
-
A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase.Mol Cell Proteomics. 2005 Jan;4(1):12-34. doi: 10.1074/mcp.M400079-MCP200. Epub 2004 Oct 25. Mol Cell Proteomics. 2005. PMID: 15507458
-
Unraveling the phosphoproteome dynamics in mammal mitochondria from a network perspective.J Proteome Res. 2013 Oct 4;12(10):4257-67. doi: 10.1021/pr4003917. Epub 2013 Sep 6. J Proteome Res. 2013. PMID: 23964737 Review.
-
Proteomics of human mitochondria.Mitochondrion. 2017 Mar;33:2-14. doi: 10.1016/j.mito.2016.07.006. Epub 2016 Jul 19. Mitochondrion. 2017. PMID: 27444749 Review.
Cited by
-
Post-translational palmitoylation of metabolic proteins.Front Physiol. 2023 Feb 24;14:1122895. doi: 10.3389/fphys.2023.1122895. eCollection 2023. Front Physiol. 2023. PMID: 36909239 Free PMC article. Review.
-
TurnoveR: A Skyline External Tool for Analysis of Protein Turnover in Metabolic Labeling Studies.J Proteome Res. 2023 Feb 3;22(2):311-322. doi: 10.1021/acs.jproteome.2c00173. Epub 2022 Sep 27. J Proteome Res. 2023. PMID: 36165806 Free PMC article.
-
MicroRNA-574 regulates FAM210A expression and influences pathological cardiac remodeling.EMBO Mol Med. 2021 Feb 5;13(2):e12710. doi: 10.15252/emmm.202012710. Epub 2020 Dec 28. EMBO Mol Med. 2021. PMID: 33369227 Free PMC article.
-
The Mitochondrial Proteomic Signatures of Human Skeletal Muscle Linked to Insulin Resistance.Int J Mol Sci. 2020 Jul 28;21(15):5374. doi: 10.3390/ijms21155374. Int J Mol Sci. 2020. PMID: 32731645 Free PMC article. Review.
-
Mitochondrial proteins: from biogenesis to functional networks.Nat Rev Mol Cell Biol. 2019 May;20(5):267-284. doi: 10.1038/s41580-018-0092-0. Nat Rev Mol Cell Biol. 2019. PMID: 30626975 Free PMC article. Review.
References
-
- Taylor SW, Fahy E, Ghosh SS. Global organellar proteomics. Trends Biotechnol. 2003;21:82–88. - PubMed
-
- Mayr M, Zhang J, Greene AS, Gutterman DD, Perloff JK, Ping P. Proteomic based development of biomarkers in cardiovascular disease: Mechanistic, clinical, and therapeutic insights. Mol Cell Proteomics. 2006;5:1853–1864. - PubMed
-
- Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol. 2005;6:702–714. - PubMed
-
- McDonald TG, Van Eyk JE. Mitochondrial proteomics. Undercover in the lipid bilayer. Basic Res Cardiol. 2003;98:219–227. - PubMed
-
- Weiss JN, Korge P, Honda HM, Ping P. Role of the mitochondrial permeability transition in myocardial disease. Circ Res. 2003;93:292–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL063901-09/HL/NHLBI NIH HHS/United States
- R01 HL065431-09/HL/NHLBI NIH HHS/United States
- HL-80111/HL/NHLBI NIH HHS/United States
- P01 HL080111/HL/NHLBI NIH HHS/United States
- R01 HL063901/HL/NHLBI NIH HHS/United States
- R01 HL065431/HL/NHLBI NIH HHS/United States
- P01 HL080111-04/HL/NHLBI NIH HHS/United States
- HL-65431/HL/NHLBI NIH HHS/United States
- HL-63901/HL/NHLBI NIH HHS/United States
- S10 RR022371/RR/NCRR NIH HHS/United States
- HL-76526/HL/NHLBI NIH HHS/United States
- RR-022371-01/RR/NCRR NIH HHS/United States
- F32 HL078109/HL/NHLBI NIH HHS/United States
- HL-78109/HL/NHLBI NIH HHS/United States
- R21 HL076526/HL/NHLBI NIH HHS/United States
- R37 HL063901/HL/NHLBI NIH HHS/United States
- R01 HL101228/HL/NHLBI NIH HHS/United States
- F32 HL078109-01/HL/NHLBI NIH HHS/United States
- R21 HL076526-02/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources


