De novo computational design of retro-aldol enzymes
- PMID: 18323453
- PMCID: PMC3431203
- DOI: 10.1126/science.1152692
De novo computational design of retro-aldol enzymes
Abstract
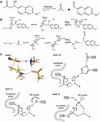
The creation of enzymes capable of catalyzing any desired chemical reaction is a grand challenge for computational protein design. Using new algorithms that rely on hashing techniques to construct active sites for multistep reactions, we designed retro-aldolases that use four different catalytic motifs to catalyze the breaking of a carbon-carbon bond in a nonnatural substrate. Of the 72 designs that were experimentally characterized, 32, spanning a range of protein folds, had detectable retro-aldolase activity. Designs that used an explicit water molecule to mediate proton shuffling were significantly more successful, with rate accelerations of up to four orders of magnitude and multiple turnovers, than those involving charged side-chain networks. The atomic accuracy of the design process was confirmed by the x-ray crystal structure of active designs embedded in two protein scaffolds, both of which were nearly superimposable on the design model.
Figures



Comment in
-
Computational design of enzymes.Chem Biol. 2008 May;15(5):421-3. doi: 10.1016/j.chembiol.2008.04.007. Chem Biol. 2008. PMID: 18482694
Similar articles
-
Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction.Science. 2010 Jul 16;329(5989):309-13. doi: 10.1126/science.1190239. Science. 2010. PMID: 20647463 Free PMC article.
-
Improving upon nature: active site remodeling produces highly efficient aldolase activity toward hydrophobic electrophilic substrates.Biochemistry. 2012 Feb 28;51(8):1658-68. doi: 10.1021/bi201899b. Epub 2012 Feb 16. Biochemistry. 2012. PMID: 22316217 Free PMC article.
-
Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli.Structure. 2001 Jan 10;9(1):1-9. doi: 10.1016/s0969-2126(00)00555-4. Structure. 2001. PMID: 11342129
-
Engineering aldolases as biocatalysts.Curr Opin Chem Biol. 2014 Apr;19(100):25-33. doi: 10.1016/j.cbpa.2013.12.010. Epub 2014 Jan 4. Curr Opin Chem Biol. 2014. PMID: 24780276 Free PMC article. Review.
-
Development of aldolase-based catalysts for the synthesis of organic chemicals.Trends Biotechnol. 2022 Mar;40(3):306-319. doi: 10.1016/j.tibtech.2021.08.001. Epub 2021 Aug 27. Trends Biotechnol. 2022. PMID: 34462144 Review.
Cited by
-
Computational design of serine hydrolases.bioRxiv [Preprint]. 2024 Aug 30:2024.08.29.610411. doi: 10.1101/2024.08.29.610411. bioRxiv. 2024. PMID: 39257749 Free PMC article. Preprint.
-
Conformational Modulation of a Mobile Loop Controls Catalysis in the (βα)8-Barrel Enzyme of Histidine Biosynthesis HisF.JACS Au. 2024 Aug 15;4(8):3258-3276. doi: 10.1021/jacsau.4c00558. eCollection 2024 Aug 26. JACS Au. 2024. PMID: 39211614 Free PMC article.
-
Substrate Turnover Dynamics Guide Ketol-Acid Reductoisomerase Redesign for Increased Specific Activity.ACS Catal. 2024 Jun 26;14(14):10491-10509. doi: 10.1021/acscatal.4c01446. eCollection 2024 Jul 19. ACS Catal. 2024. PMID: 39050899 Free PMC article.
-
Spiers Memorial Lecture: Engineering biocatalysts.Faraday Discuss. 2024 Sep 11;252(0):9-28. doi: 10.1039/d4fd00139g. Faraday Discuss. 2024. PMID: 39046423 Free PMC article.
-
Automated in vivo enzyme engineering accelerates biocatalyst optimization.Nat Commun. 2024 Apr 24;15(1):3447. doi: 10.1038/s41467-024-46574-4. Nat Commun. 2024. PMID: 38658554 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


