The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri
- PMID: 18159241
- PMCID: PMC2147049
- DOI: 10.1371/journal.pone.0001358
The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri
Abstract
Background: Arcobacter butzleri is a member of the epsilon subdivision of the Proteobacteria and a close taxonomic relative of established pathogens, such as Campylobacter jejuni and Helicobacter pylori. Here we present the complete genome sequence of the human clinical isolate, A. butzleri strain RM4018.
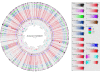
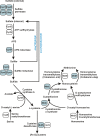
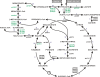
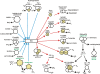
Methodology/principal findings: Arcobacter butzleri is a member of the Campylobacteraceae, but the majority of its proteome is most similar to those of Sulfuromonas denitrificans and Wolinella succinogenes, both members of the Helicobacteraceae, and those of the deep-sea vent Epsilonproteobacteria Sulfurovum and Nitratiruptor. In addition, many of the genes and pathways described here, e.g. those involved in signal transduction and sulfur metabolism, have been identified previously within the epsilon subdivision only in S. denitrificans, W. succinogenes, Sulfurovum, and/or Nitratiruptor, or are unique to the subdivision. In addition, the analyses indicated also that a substantial proportion of the A. butzleri genome is devoted to growth and survival under diverse environmental conditions, with a large number of respiration-associated proteins, signal transduction and chemotaxis proteins and proteins involved in DNA repair and adaptation. To investigate the genomic diversity of A. butzleri strains, we constructed an A. butzleri DNA microarray comprising 2238 genes from strain RM4018. Comparative genomic indexing analysis of 12 additional A. butzleri strains identified both the core genes of A. butzleri and intraspecies hypervariable regions, where <70% of the genes were present in at least two strains.
Conclusion/significance: The presence of pathways and loci associated often with non-host-associated organisms, as well as genes associated with virulence, suggests that A. butzleri is a free-living, water-borne organism that might be classified rightfully as an emerging pathogen. The genome sequence and analyses presented in this study are an important first step in understanding the physiology and genetics of this organism, which constitutes a bridge between the environment and mammalian hosts.
Conflict of interest statement
Figures
Similar articles
-
Virulence and antibiotic resistance plasticity of Arcobacter butzleri: Insights on the genomic diversity of an emerging human pathogen.Infect Genet Evol. 2020 Jun;80:104213. doi: 10.1016/j.meegid.2020.104213. Epub 2020 Jan 30. Infect Genet Evol. 2020. PMID: 32006709
-
Characterization of the emerging zoonotic pathogen Arcobacter thereius by whole genome sequencing and comparative genomics.PLoS One. 2017 Jul 3;12(7):e0180493. doi: 10.1371/journal.pone.0180493. eCollection 2017. PLoS One. 2017. PMID: 28671965 Free PMC article.
-
Genome mapping of Arcobacter butzleri.FEMS Microbiol Lett. 2006 Mar;256(2):290-7. doi: 10.1111/j.1574-6968.2006.00135.x. FEMS Microbiol Lett. 2006. PMID: 16499619
-
Insights in the pathogenesis and resistance of Arcobacter: A review.Crit Rev Microbiol. 2016 May;42(3):364-83. doi: 10.3109/1040841X.2014.954523. Epub 2015 Mar 25. Crit Rev Microbiol. 2016. PMID: 25806423 Review.
-
Arcobacter butzleri: Up-to-date taxonomy, ecology, and pathogenicity of an emerging pathogen.Compr Rev Food Sci Food Saf. 2020 Jul;19(4):2071-2109. doi: 10.1111/1541-4337.12577. Epub 2020 Jun 11. Compr Rev Food Sci Food Saf. 2020. PMID: 33337088 Review.
Cited by
-
CemR atypical response regulator impacts energy conversion in Campylobacteria.mSystems. 2024 Aug 20;9(8):e0078424. doi: 10.1128/msystems.00784-24. Epub 2024 Jul 9. mSystems. 2024. PMID: 38980050 Free PMC article.
-
Amoebae as training grounds for microbial pathogens.mBio. 2024 Aug 14;15(8):e0082724. doi: 10.1128/mbio.00827-24. Epub 2024 Jul 8. mBio. 2024. PMID: 38975782 Free PMC article. Review.
-
Evaluation of prokaryotic and eukaryotic microbial communities on microplastic-associated biofilms in marine and freshwater environments.Eng Life Sci. 2024 Mar 8;24(6):2300249. doi: 10.1002/elsc.202300249. eCollection 2024 Jun. Eng Life Sci. 2024. PMID: 38845816 Free PMC article.
-
Investigation and characterization of Aliarcobacter spp. isolated from cattle slaughterhouse in Türkiye.Int Microbiol. 2024 Aug;27(4):1321-1332. doi: 10.1007/s10123-023-00478-3. Epub 2024 Jan 11. Int Microbiol. 2024. PMID: 38206523
-
Pathogenicity assessment of Arcobacter butzleri isolated from Canadian agricultural surface water.BMC Microbiol. 2024 Jan 8;24(1):17. doi: 10.1186/s12866-023-03119-x. BMC Microbiol. 2024. PMID: 38191309 Free PMC article.
References
-
- Skirrow MB, Blaser MJ. Clinical aspects of Campylobacter infection. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ed. ed. Washington, DC: ASM Press; 2000. pp. 69–88.
-
- Marshall B. One hundred years of discovery and rediscovery of Helicobacter pylori and its association with peptic ulcer disease. In: Mobley HL, Mendz GL, Hazell SL, editors. Helicobacter pylori: Physiology and genetics. Washington, DC: ASM Press; 2001. pp. 19–24. - PubMed
-
- Sandstedt K, Ursing J, Walder M. Thermotolerant Campylobacter with no or weak catalase activity isolated from dogs. Curr Microbiol. 1983;8:209–213.
-
- Vandamme P. Taxonomy of the Family Campylobacteraceae. . In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2000. pp. 3–26.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases


