Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins
- PMID: 18042681
- PMCID: PMC2144586
- DOI: 10.1110/ps.073122908
Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins
Abstract
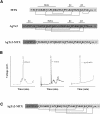
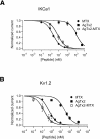
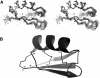
Agitoxin 2 (AgTx2) is a 38-residue scorpion toxin, cross-linked by three disulfide bridges, which acts on voltage-gated K(+) (Kv) channels. Maurotoxin (MTX) is a 34-residue scorpion toxin with an uncommon four-disulfide bridge reticulation, acting on both Ca(2+)-activated and Kv channels. A 39-mer chimeric peptide, named AgTx2-MTX, was designed from the sequence of the two toxins and chemically synthesized. It encompasses residues 1-5 of AgTx2, followed by the complete sequence of MTX. As established by enzyme cleavage, the new AgTx2-MTX molecule displays half-cystine pairings of the type C1-C5, C2-C6, C3-C7, and C4-C8, which is different from that of MTX. The 3D structure of AgTx2-MTX solved by (1)H-NMR, revealed both alpha-helical and beta-sheet structures, consistent with a common alpha/beta scaffold of scorpion toxins. Pharmacological assays of AgTx2-MTX revealed that this new molecule is more potent than both original toxins in blocking rat Kv1.2 channel. Docking simulations, performed with the 3D structure of AgTx2-MTX, confirmed this result and demonstrated the participation of the N-terminal domain of AgTx2 in its increased affinity for Kv1.2 through additional molecular contacts. Altogether, the data indicated that replacement of the N-terminal domain of MTX by the one of AgTx2 in the AgTx2-MTX chimera results in a reorganization of the disulfide bridge arrangement and an increase of affinity to the Kv1.2 channel.
Figures

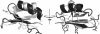
Similar articles
-
Synthesis, 1H NMR structure, and activity of a three-disulfide-bridged maurotoxin analog designed to restore the consensus motif of scorpion toxins.J Biol Chem. 2000 May 5;275(18):13605-12. doi: 10.1074/jbc.275.18.13605. J Biol Chem. 2000. PMID: 10788477
-
Synthesis, 3-D structure, and pharmacology of a reticulated chimeric peptide derived from maurotoxin and Tsk scorpion toxins.Biochem Biophys Res Commun. 2002 Mar 1;291(3):640-8. doi: 10.1006/bbrc.2002.6496. Biochem Biophys Res Commun. 2002. PMID: 11855838
-
Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins.J Biol Chem. 2004 Dec 31;279(53):55690-6. doi: 10.1074/jbc.M410055200. Epub 2004 Oct 21. J Biol Chem. 2004. PMID: 15498765
-
Disulfide bridge reorganization induced by proline mutations in maurotoxin.FEBS Lett. 2001 Feb 2;489(2-3):202-7. doi: 10.1016/s0014-5793(00)02433-9. FEBS Lett. 2001. PMID: 11165250
-
Toxin bioportides: exploring toxin biological activity and multifunctionality.Cell Mol Life Sci. 2017 Feb;74(4):647-661. doi: 10.1007/s00018-016-2343-6. Epub 2016 Aug 23. Cell Mol Life Sci. 2017. PMID: 27554773 Free PMC article. Review.
Cited by
-
AgTx2-GFP, Fluorescent Blocker Targeting Pharmacologically Important Kv1.x (x = 1, 3, 6) Channels.Toxins (Basel). 2023 Mar 18;15(3):229. doi: 10.3390/toxins15030229. Toxins (Basel). 2023. PMID: 36977120 Free PMC article.
-
Kv1.3 Channel as a Key Therapeutic Target for Neuroinflammatory Diseases: State of the Art and Beyond.Front Neurosci. 2020 Jan 14;13:1393. doi: 10.3389/fnins.2019.01393. eCollection 2019. Front Neurosci. 2020. PMID: 31992966 Free PMC article. Review.
-
Molecular Dynamics Simulation Reveals Specific Interaction Sites between Scorpion Toxins and Kv1.2 Channel: Implications for Design of Highly Selective Drugs.Toxins (Basel). 2017 Nov 1;9(11):354. doi: 10.3390/toxins9110354. Toxins (Basel). 2017. PMID: 29104247 Free PMC article.
-
Scorpion toxins specific for potassium (K+) channels: a historical overview of peptide bioengineering.Toxins (Basel). 2012 Nov 1;4(11):1082-119. doi: 10.3390/toxins4111082. Toxins (Basel). 2012. PMID: 23202307 Free PMC article. Review.
-
Developing a comparative docking protocol for the prediction of peptide selectivity profiles: investigation of potassium channel toxins.Toxins (Basel). 2012 Feb;4(2):110-38. doi: 10.3390/toxins4020110. Epub 2012 Feb 6. Toxins (Basel). 2012. PMID: 22474570 Free PMC article.
References
-
- Bartels, C., Xia, T.H., Billeter, M., Guntert, P., Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:110–118. - PubMed
-
- Blanc, E., Sabatier, J.M., Kharrat, R., Meunier, S., El Ayeb, M., Van Reitschoten, J., Darbon, H. Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels. Proteins. 1997;29:321–333. - PubMed
-
- Bontems, F., Roumestand, C., Gilquin, B., Ménez, A., Toma, F. Refined structure of charybdotoxin: Common motifs in scorpion toxins and insect defensins. Science. 1991;254:1521–1523. - PubMed
-
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
-
- Darbon, H., Blanc, E., Sabatier, J.M. Animal toxins and potassium channels. In: Darbon H., Sabatier J.M., editors. Perspectives in drug discovery and design. Vol. 15. Kluwer/Dordrecht; The Netherlands: 1999. pp. 40–60.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

