Recent developments in yeast aging
- PMID: 17530929
- PMCID: PMC1877880
- DOI: 10.1371/journal.pgen.0030084
Recent developments in yeast aging
Abstract
In the last decade, research into the molecular determinants of aging has progressed rapidly and much of this progress can be attributed to studies in invertebrate eukaryotic model organisms. Of these, single-celled yeast is the least complicated and most amenable to genetic and molecular manipulations. Supporting the use of this organism for aging research, increasing evidence has accumulated that a subset of pathways influencing longevity in yeast are conserved in other eukaryotes, including mammals. Here we briefly outline aging in yeast and describe recent findings that continue to keep this "simple" eukaryote at the forefront of aging research.
Conflict of interest statement
Competing interests. BKK and MK have a pending patent application for technology for the identification of new aging genes. BKK has a minor stock holding in Elixir Pharmaceuticals.
Figures
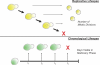
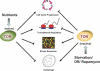
Similar articles
-
Mice share yeast's ageing system.Nature. 2008 Nov 27;456(7221):433. doi: 10.1038/456433a. Nature. 2008. PMID: 19037283 No abstract available.
-
Longevity research. Single signal unites treatments that prolong life.Science. 2003 May 9;300(5621):881-3. doi: 10.1126/science.300.5621.881a. Science. 2003. PMID: 12738822 No abstract available.
-
Does resveratrol activate yeast Sir2 in vivo?Aging Cell. 2007 Aug;6(4):415-6. doi: 10.1111/j.1474-9726.2007.00314.x. Aging Cell. 2007. PMID: 17635418 No abstract available.
-
Sound silencing: the Sir2 protein and cellular senescence.Bioessays. 2001 Apr;23(4):327-32. doi: 10.1002/bies.1047. Bioessays. 2001. PMID: 11268038 Review.
-
Small molecules that regulate lifespan: evidence for xenohormesis.Mol Microbiol. 2004 Aug;53(4):1003-9. doi: 10.1111/j.1365-2958.2004.04209.x. Mol Microbiol. 2004. PMID: 15306006 Review.
Cited by
-
Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction.Cells. 2024 Sep 1;13(17):1470. doi: 10.3390/cells13171470. Cells. 2024. PMID: 39273040 Free PMC article.
-
Delayed luminescence to monitor growth stages and assess the entropy of Saccharomyces cerevisiae.Heliyon. 2024 Mar 22;10(7):e27866. doi: 10.1016/j.heliyon.2024.e27866. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38623220 Free PMC article.
-
PICLS with human cells is the first high throughput screening method for identifying novel compounds that extend lifespan.Biol Direct. 2024 Jan 23;19(1):8. doi: 10.1186/s13062-024-00455-4. Biol Direct. 2024. PMID: 38254217 Free PMC article.
-
Cellular Stress Impact on Yeast Activity in Biotechnological Processes-A Short Overview.Microorganisms. 2023 Oct 9;11(10):2522. doi: 10.3390/microorganisms11102522. Microorganisms. 2023. PMID: 37894181 Free PMC article. Review.
-
Skeletal muscle mitochondrial interactome remodeling is linked to functional decline in aged female mice.Nat Aging. 2023 Mar;3(3):313-326. doi: 10.1038/s43587-023-00366-5. Epub 2023 Mar 2. Nat Aging. 2023. PMID: 37118428 Free PMC article.
References
-
- Jazwinski SM. Longevity, genes, and aging: A view provided by a genetic model system. Exp Gerontol. 1999;34:1–6. - PubMed
-
- Kaeberlein M. Longevity and aging in the budding yeast. In: Conn PM, editor. Handbook of models for human aging. Boston: Elvesier Press; 2006. pp. 109–120.
-
- Piper PW. Long-lived yeast as a model for ageing research. Yeast. 2006;23:215–226. - PubMed
-
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


