Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution
- PMID: 17480121
- PMCID: PMC1865561
- DOI: 10.1371/journal.pgen.0030065
Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution
Abstract
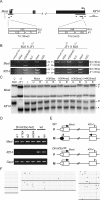
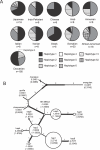
Imprinted genes are expressed in a parent-of-origin manner and are located in clusters throughout the genome. Aberrations in the expression of imprinted genes on human Chromosome 7 have been suggested to play a role in the etiologies of Russell-Silver Syndrome and autism. We describe the imprinting of KLF14, an intronless member of the Krüppel-like family of transcription factors located at Chromosome 7q32. We show that it has monoallelic maternal expression in all embryonic and extra-embryonic tissues studied, in both human and mouse. We examine epigenetic modifications in the KLF14 CpG island in both species and find this region to be hypomethylated. In addition, we perform chromatin immunoprecipitation and find that the murine Klf14 CpG island lacks allele-specific histone modifications. Despite the absence of these defining features, our analysis of Klf14 in offspring from DNA methyltransferase 3a conditional knockout mice reveals that the gene's expression is dependent upon a maternally methylated region. Due to the intronless nature of Klf14 and its homology to Klf16, we suggest that the gene is an ancient retrotransposed copy of Klf16. By sequence analysis of numerous species, we place the timing of this event after the divergence of Marsupialia, yet prior to the divergence of the Xenarthra superclade. We identify a large number of sequence variants in KLF14 and, using several measures of diversity, we determine that there is greater variability in the human lineage with a significantly increased number of nonsynonymous changes, suggesting human-specific accelerated evolution. Thus, KLF14 may be the first example of an imprinted transcript undergoing accelerated evolution in the human lineage.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


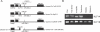
Similar articles
-
Klf14 is an imprinted transcription factor that regulates placental growth.Placenta. 2019 Dec;88:61-67. doi: 10.1016/j.placenta.2019.09.013. Epub 2019 Sep 27. Placenta. 2019. PMID: 31675530 Free PMC article.
-
Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes.Hum Mol Genet. 2011 Aug 15;20(16):3188-97. doi: 10.1093/hmg/ddr224. Epub 2011 May 18. Hum Mol Genet. 2011. PMID: 21593219
-
Organization and parent-of-origin-specific methylation of imprinted Peg3 gene on mouse proximal chromosome 7.Genomics. 2000 Feb 1;63(3):333-40. doi: 10.1006/geno.1999.6103. Genomics. 2000. PMID: 10704281
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene.Cytogenet Genome Res. 2006;113(1-4):202-8. doi: 10.1159/000090833. Cytogenet Genome Res. 2006. PMID: 16575181 Review.
Cited by
-
Enhancer of Zeste Homolog 2 Protects Mucosal Melanoma from Ferroptosis via the KLF14-SLC7A11 Signaling Pathway.Cancers (Basel). 2024 Oct 30;16(21):3660. doi: 10.3390/cancers16213660. Cancers (Basel). 2024. PMID: 39518098 Free PMC article.
-
Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms.Metabolites. 2023 Jan 29;13(2):199. doi: 10.3390/metabo13020199. Metabolites. 2023. PMID: 36837818 Free PMC article. Review.
-
KLF14 regulates the growth of hepatocellular carcinoma cells via its modulation of iron homeostasis through the repression of iron-responsive element-binding protein 2.J Exp Clin Cancer Res. 2023 Jan 5;42(1):5. doi: 10.1186/s13046-022-02562-4. J Exp Clin Cancer Res. 2023. PMID: 36600258 Free PMC article.
-
Kruppel-like Factors in Skeletal Physiology and Pathologies.Int J Mol Sci. 2022 Dec 2;23(23):15174. doi: 10.3390/ijms232315174. Int J Mol Sci. 2022. PMID: 36499521 Free PMC article. Review.
-
Differential Genetic and Epigenetic Effects of the KLF14 Gene on Body Shape Indices and Metabolic Traits.Int J Mol Sci. 2022 Apr 9;23(8):4165. doi: 10.3390/ijms23084165. Int J Mol Sci. 2022. PMID: 35456983 Free PMC article.
References
-
- Walter J, Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum Mol Genet. 2003;12(Spec No 2):R 215–R220. - PubMed
-
- Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, et al. Epimutation of the telomeric imprinting center region on Chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. - PubMed
-
- Eggermann T, Wollmann HA, Kuner R, Eggermann K, Enders H, et al. Molecular studies in 37 Silver-Russell syndrome patients: Frequency and etiology of uniparental disomy. Hum Genet. 1997;100:415–419. - PubMed
-
- Nakabayashi K, Fernandez BA, Teshima I, Shuman C, Proud VK, et al. Molecular genetic studies of human Chromosome 7 in Russell-Silver syndrome. Genomics. 2002;79:186–196. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases


