The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase
- PMID: 15838047
- PMCID: PMC1082839
- DOI: 10.1128/JB.187.9.3201-3205.2005
The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase
Abstract
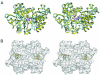
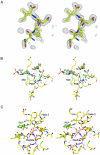
The biosynthesis of cysteine in bacteria and plants is carried out by a two-step pathway, catalyzed by serine acetyltransferase (SAT) and O-acetylserine sulfhydrylase (OASS; O-acetylserine [thiol] lyase). The aerobic form of OASS forms a tight bienzyme complex with SAT in vivo, termed cysteine synthase. We have determined the crystal structure of OASS in complex with a C-terminal peptide of SAT required for bienzyme complex formation. The binding site of the peptide is at the active site of OASS, and its C-terminal carboxyl group occupies the same anion binding pocket as the alpha-carboxylate of the O-acetylserine substrate of OASS. These results explain the partial inhibition of OASS by SAT on complex formation as well as the competitive dissociation of the complex by O-acetylserine.
Figures
Similar articles
-
Interaction of serine acetyltransferase with O-acetylserine sulfhydrylase active site: evidence from fluorescence spectroscopy.Protein Sci. 2005 Aug;14(8):2115-24. doi: 10.1110/ps.051492805. Epub 2005 Jun 29. Protein Sci. 2005. PMID: 15987896 Free PMC article.
-
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural and kinetic properties of the free and bound enzymes.Eur J Biochem. 1998 Jul 1;255(1):235-45. doi: 10.1046/j.1432-1327.1998.2550235.x. Eur J Biochem. 1998. PMID: 9692924
-
Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex.Plant Cell. 2006 Dec;18(12):3647-55. doi: 10.1105/tpc.106.047316. Epub 2006 Dec 28. Plant Cell. 2006. PMID: 17194764 Free PMC article.
-
The cysteine regulatory complex from plants and microbes: what was old is new again.Curr Opin Struct Biol. 2013 Apr;23(2):302-10. doi: 10.1016/j.sbi.2013.02.011. Epub 2013 Mar 17. Curr Opin Struct Biol. 2013. PMID: 23510784 Review.
-
Advancements in inhibitors of crucial enzymes in the cysteine biosynthetic pathway: Serine acetyltransferase and O-acetylserine sulfhydrylase.Chem Biol Drug Des. 2024 Jul;104(1):e14573. doi: 10.1111/cbdd.14573. Chem Biol Drug Des. 2024. PMID: 38965664 Review.
Cited by
-
Combatting antimicrobial resistance via the cysteine biosynthesis pathway in bacterial pathogens.Biosci Rep. 2022 Oct 28;42(10):BSR20220368. doi: 10.1042/BSR20220368. Biosci Rep. 2022. PMID: 36148777 Free PMC article. Review.
-
Inhibitors of O-Acetylserine Sulfhydrylase with a Cyclopropane-Carboxylic Acid Scaffold Are Effective Colistin Adjuvants in Gram Negative Bacteria.Pharmaceuticals (Basel). 2022 Jun 20;15(6):766. doi: 10.3390/ph15060766. Pharmaceuticals (Basel). 2022. PMID: 35745685 Free PMC article.
-
New insights into the structure and function of an emerging drug target CysE.3 Biotech. 2021 Aug;11(8):373. doi: 10.1007/s13205-021-02891-9. Epub 2021 Jul 18. 3 Biotech. 2021. PMID: 34367865 Free PMC article. Review.
-
A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain.Nat Commun. 2021 Mar 2;12(1):1392. doi: 10.1038/s41467-021-21282-5. Nat Commun. 2021. PMID: 33654102 Free PMC article.
-
Molecular mechanism of selective substrate engagement and inhibitor disengagement of cysteine synthase.J Biol Chem. 2021 Jan-Jun;296:100041. doi: 10.1074/jbc.RA120.014490. Epub 2020 Nov 24. J Biol Chem. 2021. PMID: 33162395 Free PMC article.
References
-
- Becker, M. A., N. M. Kredich, and G. M. Tomkins. 1969. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J. Biol. Chem. 244:2418-2427. - PubMed
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. - PubMed
-
- Burkhard, P., G. S. Rao, E. Hohenester, K. D. Schnackerz, P. F. Cook, and J. N. Jansonius. 1998. Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol. 283:121-133. - PubMed
-
- Burkhard, P., C. H. Tai, J. N. Jansonius, and P. F. Cook. 2000. Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: structure of the enzyme with chloride bound. J. Mol. Biol. 303:279-286. - PubMed
-
- Burkhard, P., C. H. Tai, C. M. Ristroph, P. F. Cook, and J. N. Jansonius. 1999. Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol. 291:941-953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


